
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,963,510 – 17,963,666 |
| Length | 156 |
| Max. P | 0.874873 |

| Location | 17,963,510 – 17,963,666 |
|---|---|
| Length | 156 |
| Sequences | 6 |
| Columns | 165 |
| Reading direction | reverse |
| Mean pairwise identity | 54.33 |
| Shannon entropy | 0.85547 |
| G+C content | 0.33319 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -4.63 |
| Energy contribution | -6.47 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.16 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
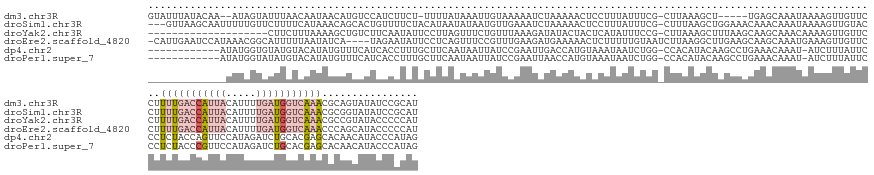
>dm3.chr3R 17963510 156 - 27905053 GUAUUUAUACAA--AUAGUAUUUAACAAUAACAUGUCCAUCUUCU-UUUUAUAAAUUGUAAAAAUCUAAAAACUCCUUUAUUUCG-CUUAAAGCU-----UGAGCAAAUAAAAGUUGUUCCUUUUGACCAUUACAUUUUGAUGGUCAAACGCAGUAUAUCCGCAU ((...(((((..--............((((((............(-((((((.....)))))))............(((((((.(-((((.....-----)))))))))))).))))))...(((((((((((.....)))))))))))....)))))...)).. ( -29.00, z-score = -3.38, R) >droSim1.chr3R 17779762 161 - 27517382 ---GUUAAGCAAUUUUUGUUCUUUUCAUAAACAGCACUGUUUUCUACAUAAUAUAAUGUUGAAAUCUAAAAACUCCUUUAUUUCG-CUUUAAGCUGGAAACAAACAAAUAAAAGUUGUACCUUUUGACCAUUACAUUUUGAUGGUCAAACGCGGUAUAUCCGCAU ---.....((((((((((((..(((......((((.....((((.((((......)))).))))..((((......)))).....-......)))).....)))..))))))))))))....(((((((((((.....))))))))))).((((.....)))).. ( -37.40, z-score = -4.11, R) >droYak2.chr3R 18812089 144 - 28832112 --------------------CUUCUUUAAAAGCUGUCUUCAAUAUUCCUUAGUUUCUGUUUAAAGAUAUACUACUCAUAUUUCCG-CUUAAAGCUUUAAGCAAGCAAACAAAAGUUGUUCCUUUUGACCAUUACAUUUUGAUGGUCAAACGCCGUAUACCCCCAU --------------------..(((((((((((((..............)))))....))))))))(((((.............(-(((((....))))))..((.((((.....))))...(((((((((((.....))))))))))).)).)))))....... ( -27.64, z-score = -3.32, R) >droEre2.scaffold_4820 355655 160 + 10470090 -CAUUGAAUCCAUAAACGGCAUUUUUAAUAUCA----UAGAAUAUUCCUCAGUUUCCGUUUGAAGAUGAAAAACUCUUUUUGUAAUCUUAAGGCUUGAAGCAAGCAAAUGAAAGUUGUUCCUUUUGACCAUUACAUUUUGAUGGUCAAACCCAGCAUACCCCCAU -((((.......(((((((.......(((((..----....))))).........)))))))(((((..((((....))))...)))))...(((((...))))).))))...((((.....(((((((((((.....)))))))))))..)))).......... ( -28.49, z-score = -0.53, R) >dp4.chr2 1878717 151 - 30794189 ------------AUAUGGUGUAUGUACAUAUGUUUCAUCACCUUUGCUUCAAUAAUUAUCCGAAUUGACCAUGUAAAUAAUCUGG-CCACAUACAAGCCUGAAACAAAU-AUCUUUAUUCCCUCUACCAGUUCCAUAGAUCUGCACGAGCACAACAUACCCAUAG ------------.(((((.((((((.....(((((((.....(((((.(((((..........)))))....)))))......((-(.........))))))))))...-...........(((...(((.((....)).)))...)))....))))))))))). ( -28.90, z-score = -2.24, R) >droPer1.super_7 2042164 151 - 4445127 ------------AUAUGGUAUAUGUACAUAUGUUUCAUCACCUUUGCUUCAAUAAUUAUCCGAAUUAACCAUGUAAAUAAUCUGG-CCACAUACAAGCCUGAAACAAAU-AUCUUUAUUCCCUCUACCCGUUCCAUAGAUCUGCACGAGCACAACAUACCCAUAG ------------.(((((..(((((.....(((((((.....(((((.....(((((.....))))).....)))))......((-(.........))))))))))...-...................((((.............))))...))))).))))). ( -20.82, z-score = -0.70, R) >consensus _____________UAUGGUAUUUGUAAAUAACAUGCCUAACAUAUGCUUCAAUAAUUGUUCAAAUAUAAAAAAUACAUAAUUUCG_CUUAAAGCUAGAAGCAAACAAAUAAAAGUUGUUCCUUUUGACCAUUACAUUUUGAUGGUCAAACGCAGCAUACCCACAU ..........................................................................................................................(((((((((((.....)))))))))))................ ( -4.63 = -6.47 + 1.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:20 2011