| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,960,285 – 17,960,344 |
| Length | 59 |
| Max. P | 0.500000 |

| Location | 17,960,285 – 17,960,344 |
|---|---|
| Length | 59 |
| Sequences | 9 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 77.90 |
| Shannon entropy | 0.41501 |
| G+C content | 0.44785 |
| Mean single sequence MFE | -13.69 |
| Consensus MFE | -8.59 |
| Energy contribution | -8.17 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
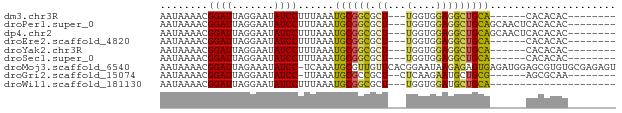
Download alignment: ClustalW | MAF
>dm3.chr3R 17960285 59 - 27905053 AAUAAAACGGAUUAGGAAUAUCCUUUAAAUGCGGCGCU---UGGUGGAGGCUGCA------CACACAC-------- ........((((.......))))......((((((.((---(....)))))))))------.......-------- ( -13.50, z-score = -1.55, R) >droPer1.super_0 3455095 65 + 11822988 AAUAAAACGGAUUAGGAAUAUCCUUUAAAUGCGGCGCU---UGGUGGAGGCUGCAGCAACUCACACAC-------- ........((((.......))))......((((((.((---(....))))))))).............-------- ( -13.80, z-score = -0.69, R) >dp4.chr2 12129116 65 - 30794189 AAUAAAACGGAUUAGGAAUAUCCUUUAAAUGCGGCGCU---UGGUGGAGGCUGCAGCAACUCACACAC-------- ........((((.......))))......((((((.((---(....))))))))).............-------- ( -13.80, z-score = -0.69, R) >droEre2.scaffold_4820 352356 59 + 10470090 AAUAAAACGGAUUAGGAAUAUCCUUUAAAUGCGGCGCU---UGGUGGAGGCUGCA------CACACAC-------- ........((((.......))))......((((((.((---(....)))))))))------.......-------- ( -13.50, z-score = -1.55, R) >droYak2.chr3R 18808683 59 - 28832112 AAUAAAACGGAUUAGGAAUAUCCUUUAAAUGCGGCGCU---UGGUGGAGGCUGCA------CACACAC-------- ........((((.......))))......((((((.((---(....)))))))))------.......-------- ( -13.50, z-score = -1.55, R) >droSec1.super_0 18330616 59 - 21120651 AAUAAAACGGAUUAGGAAUAUCCUUUAAAUGCGGCGCU---UGGUGGAGGCUGCA------CACACAC-------- ........((((.......))))......((((((.((---(....)))))))))------.......-------- ( -13.50, z-score = -1.55, R) >droMoj3.scaffold_6540 27639001 75 - 34148556 AAUAAAACGGAUUAGAAAUAUCC-UCAAAUGCGUUGUUCACGGAAUAAGAGAGUGAGAUGGAGCGUGUGCGAGAGU ........((((.(....)))))-((..(((((((.(((((...........)))))....)))))))..)).... ( -13.20, z-score = -0.30, R) >droGri2.scaffold_15074 1096883 59 - 7742996 AAUAAAACGGAUUAGGAAUAUCC-UUAAAUGCGCCGCU--CUCAAGAAUGCUGCG------AGCGCAA-------- ........((((.......))))-.....((((((((.--............)))------.))))).-------- ( -15.62, z-score = -1.49, R) >droWil1.scaffold_181130 12101917 52 - 16660200 AAUAAAACGGAUUAGGAAUAUCCUUUAAAUGCGGCGCU---UGGUGGAUGCUGCA--------------------- ........((((.......))))......(((((((((---....)).)))))))--------------------- ( -12.80, z-score = -1.32, R) >consensus AAUAAAACGGAUUAGGAAUAUCCUUUAAAUGCGGCGCU___UGGUGGAGGCUGCA______CACACAC________ ........((((.......))))......((((((..............))))))..................... ( -8.59 = -8.17 + -0.42)
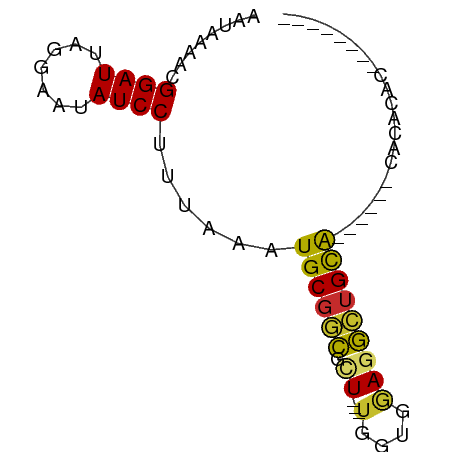

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:18 2011