
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,938,557 – 17,938,657 |
| Length | 100 |
| Max. P | 0.634398 |

| Location | 17,938,557 – 17,938,657 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.45858 |
| G+C content | 0.44455 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -12.16 |
| Energy contribution | -13.44 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17938557 100 + 27905053 GACAAUUUGCCGCCUGAUUAAUGAGUGUCUGUGGUUCUCCGCCAGACAAAACCAGCGGAAUGCCAGUUG---GCCAGAUGCGGGAUUAUUUGCGGUAAUAUUU ......(((((((.(((((......(((((((((....))).))))))...((.(((....(((....)---))....))))))))))...)))))))..... ( -31.30, z-score = -1.47, R) >droSim1.chr3R 17744940 100 + 27517382 GACAAUUUGCCGCCUGAUUAAUGAGUGUCUGUGGUUCUCCGCCAGACAAAACCAGCGGAAUGCCAGUUG---GCCAGAUGCGGGAUUAUUUGCGGUAAUAUUU ......(((((((.(((((......(((((((((....))).))))))...((.(((....(((....)---))....))))))))))...)))))))..... ( -31.30, z-score = -1.47, R) >droSec1.super_0 18307081 100 + 21120651 GACAAUUUGCCGCCUGAUUAAUGAGUGUCUGUGGUUCUCCGCCAGACAAAACCAGCGGAAUGCCAGUUG---GCCAGAUGCGGGAUUAUUUGCGGUAAUAUUU ......(((((((.(((((......(((((((((....))).))))))...((.(((....(((....)---))....))))))))))...)))))))..... ( -31.30, z-score = -1.47, R) >droYak2.chr3R 18782769 94 + 28832112 GACAAUUUGCCGCCUGAUUAAUGAGUGUCUGUGGUUC------AGACAAAACCAGCGGAAUGCCAGUUG---GGCAGAUGCGAGAUUAUUUGCGGUCAUAUUU (((.(((((((..............((((((.....)------)))))....((((((....)).))))---)))))))(((((....))))).)))...... ( -27.10, z-score = -1.16, R) >droEre2.scaffold_4820 331894 100 - 10470090 GACAAUUUGCCGCCUGAUUAAUGAGUGUCUGUGGUUCUCCGCCAGACAAAACCAGCGGAAUGACAGUUG---GGCAGAUGCGAGAUUAUUUGCGGUAAUAUUU ......(((((((..(((......(((((((((((.....)))........(((((.........))))---)))))))))......))).)))))))..... ( -29.30, z-score = -1.51, R) >droAna3.scaffold_13340 8944351 102 + 23697760 GACAAUUUGCCGCCUGAUUAAUGACUGUCUGUGGCUCUCCACCAAA-AAAUAGAGAGUUAAGCCAGCAGACUGGCAGAUGUGAGAUUAUUUGCGGCAAUAUUU ......(((((((..(((...(.((.((((.((((((((.......-.....)))))))).(((((....))))))))))).)....))).)))))))..... ( -31.90, z-score = -3.00, R) >dp4.chr2 12104009 94 + 30794189 GACAAUUUGCCGCCUGAUUAAUGACCGUCUGUGGCCAUCCACCAGACACAAUAGACACA----CACACA-----CAGAUGUGAGAUUAUUUGCGGUAAUAUUU ......(((((((.(((((.......((((((((....))).)))))............----((((..-----....)))).)))))...)))))))..... ( -23.20, z-score = -2.44, R) >droPer1.super_0 3430049 92 - 11822988 GACAAUUUGCCGCCUGAUUAAUGACCGUCUGUGGCCAUCCACCAGACACAAUAGACA------CACACA-----CAGAUGUGAGAUUAUUUGCGGUAAUAUUU ......(((((((.(((((.......((((((((....))).)))))..........------((((..-----....)))).)))))...)))))))..... ( -23.20, z-score = -2.56, R) >droWil1.scaffold_181130 12080119 85 + 16660200 GACAAUUUACCGCCUGAUUAAUGACAGUCUGUGGUUCUUUUU-------GCCCAGA-------CAGACA----CAAAUUGUAUUAUUAUAUACCAUAAUAUUG .(((((((...(.(........).).(((((.(((.......-------)))))))-------).....----.)))))))..((((((.....))))))... ( -14.20, z-score = -1.43, R) >consensus GACAAUUUGCCGCCUGAUUAAUGAGUGUCUGUGGUUCUCCGCCAGACAAAACCAGCGGAAUGCCAGUUG___GCCAGAUGCGAGAUUAUUUGCGGUAAUAUUU ......(((((((.............((((((((....)))).))))............................(((((......))))))))))))..... (-12.16 = -13.44 + 1.29)

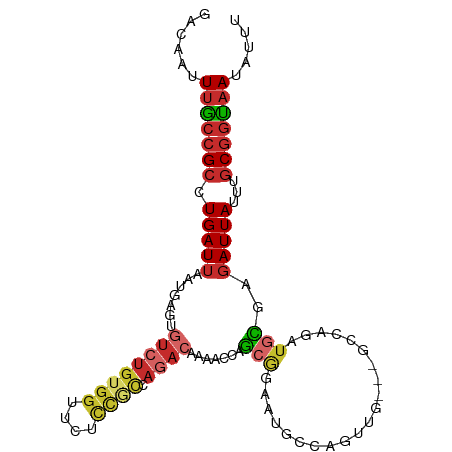
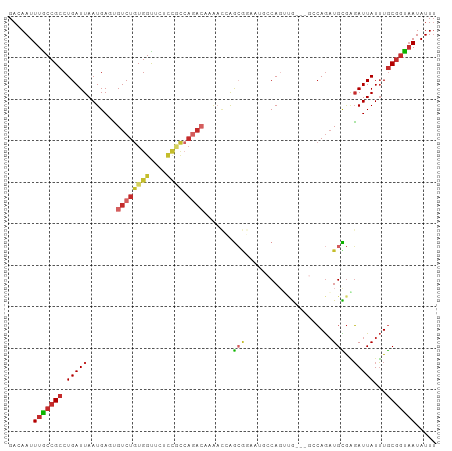
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:13 2011