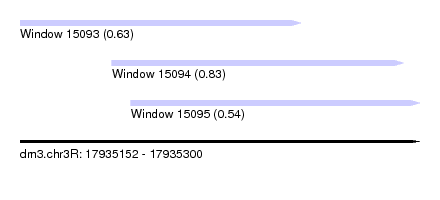
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,935,152 – 17,935,300 |
| Length | 148 |
| Max. P | 0.832351 |

| Location | 17,935,152 – 17,935,256 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.45 |
| Shannon entropy | 0.40816 |
| G+C content | 0.50390 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17935152 104 + 27905053 UCUCUGUUUCACCAACCAACCAGGCAUUCAUCCAUCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGG ...........(((..((((..((((((.......((..((((....))))..)).......))))))......((((((((....))))))))...))))))) ( -25.34, z-score = -1.17, R) >droSim1.chr3R 17740953 104 + 27517382 UCUCUGUUCCACCAACCAACCAGGCAUCCAUCCAUCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGG ........((((.(((......(((((........((..((((....))))..))........)))))......((((((((....))))))))...))))))) ( -25.59, z-score = -1.13, R) >droSec1.super_0 18303675 104 + 21120651 UCUCUGUUCCACCAACCAAGCAGGCAUCCAUCCAUCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGG ........((((.(((...((((((((........((..((((....))))..))........))))..)))).((((((((....))))))))...))))))) ( -28.39, z-score = -1.77, R) >droYak2.chr3R 18779215 104 + 28832112 UCUCUCUUCCACCAACCAGCCUUGCAGCCAUCCGUCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGG ........((((.(((......(((((......(.((..((((....))))..)).)(((....)))..)))))((((((((....))))))))...))))))) ( -27.40, z-score = -1.66, R) >droEre2.scaffold_4820 328620 96 - 10470090 UCUCUGUUCCACCAGCCAG--------CCAUCCAUCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGG ........((((.(((..(--------(...........((((....))))..(((.(((....))).))))).((((((((....))))))))...))))))) ( -22.40, z-score = -0.27, R) >droAna3.scaffold_13340 8941204 94 + 23697760 -GCCUCCUCCUCCAGCCG---------CCGCAUGCCUCCCCUUCCUCUCAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGG -..........(((...(---------((((((((..................(((.(((....))).))))))))))))(((((((.....)))))))..))) ( -24.06, z-score = -1.72, R) >dp4.chr2 12099515 91 + 30794189 ---CUCUGUCUCUGUUCCG---------CACGGGUC-CUGUGUCGGUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGG ---..(((.(((....(((---------(((((...-))))).)))..))).)))............(((.(((((((((((....))))))))..))).))). ( -26.90, z-score = -0.96, R) >droPer1.super_0 3425723 86 - 11822988 --------UCUCUGUCCCA---------CACUGGUC-CUGUGUCGGUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGG --------....(((.(((---------.(((.(.(-(......)).).))))))..))).......(((.(((((((((((....))))))))..))).))). ( -25.50, z-score = -0.81, R) >consensus UCUCUGUUCCACCAACCAA________CCAUCCAUCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGG ...........................................(((...((((((.((((....((((((.....))))))....)))).).)))))...))). (-17.36 = -17.50 + 0.14)



| Location | 17,935,186 – 17,935,294 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.86 |
| Shannon entropy | 0.29243 |
| G+C content | 0.44345 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -23.31 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.832351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17935186 108 + 27905053 UCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA-UG- .....((((....)))).((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).......-..- ( -28.00, z-score = -1.39, R) >droSim1.chr3R 17740987 108 + 27517382 UCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA-UG- .....((((....)))).((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).......-..- ( -28.00, z-score = -1.39, R) >droSec1.super_0 18303709 108 + 21120651 UCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA-UG- .....((((....)))).((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).......-..- ( -28.00, z-score = -1.39, R) >droYak2.chr3R 18779249 108 + 28832112 UCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCCAUCUUCAA-UG- .....((((....)))).((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).......-..- ( -30.30, z-score = -1.90, R) >droEre2.scaffold_4820 328646 108 - 10470090 UCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA-UG- .....((((....)))).((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).......-..- ( -28.00, z-score = -1.39, R) >droAna3.scaffold_13340 8941235 101 + 23697760 -------UUCCUCUCAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGCCGGGUAUCUCCAGUU-- -------.(((...(((((((.((((....((((((.....))))))....)))).).))))))((((((((........))))))))......)))...........-- ( -28.00, z-score = -1.56, R) >dp4.chr2 12099545 100 + 30794189 ---------CGGUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCAAAUAAACACGUCAUAAACCGUUGACUAUCUUCAAGUG- ---------.....(((.((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).)))......- ( -28.80, z-score = -1.66, R) >droPer1.super_0 3425748 101 - 11822988 ---------CGGUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCAAAUAAACCCGUCAUAAACCGUUGACUAUCUUCAAGUGC ---------.....(((.((((((......((((((.(((((((((((....))))))))..))).))))))........((........)).)))))).)))....... ( -27.60, z-score = -1.08, R) >droWil1.scaffold_181130 12072052 84 + 16660200 -----------------UUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUU-CAGUGGUUAAUAAAUACGUCAUAAGCCGC---CCAACUCCA----- -----------------((((.(..........(((((..((((((((....))))))))...-)))))((........))..........).---)))).....----- ( -19.80, z-score = -0.93, R) >consensus UCUCUCUCGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA_UG_ ..............(((.(((((.......((((((.(((((((((((....))))))))..))).))))))........((........))..))))).)))....... (-23.31 = -23.64 + 0.34)



| Location | 17,935,193 – 17,935,300 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.77 |
| Shannon entropy | 0.28106 |
| G+C content | 0.45912 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -23.31 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17935193 107 + 27905053 CGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA-UGCCUCGU-- .(((.(.(((.((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).))).).-))).....-- ( -30.80, z-score = -1.17, R) >droSim1.chr3R 17740994 107 + 27517382 CGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA-UGCCUCGU-- .(((.(.(((.((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).))).).-))).....-- ( -30.80, z-score = -1.17, R) >droSec1.super_0 18303716 107 + 21120651 CGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA-UGCCUCGU-- .(((.(.(((.((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).))).).-))).....-- ( -30.80, z-score = -1.17, R) >droYak2.chr3R 18779256 107 + 28832112 CGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCCAUCUUCAA-UGACUCGU-- ......(((((((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).......-..))))).-- ( -31.50, z-score = -1.15, R) >droEre2.scaffold_4820 328653 107 - 10470090 CGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA-UGCCUCGU-- .(((.(.(((.((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).))).).-))).....-- ( -30.80, z-score = -1.17, R) >droAna3.scaffold_13340 8941242 100 + 23697760 -------CAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGCCGGGUAUCUCCAGUUGCCUGU--- -------(((((((.((((....((((((.....))))))....)))).).))))))((((((((........)))))))).....((((((........)))))).--- ( -32.60, z-score = -1.85, R) >dp4.chr2 12099545 106 + 30794189 --CGGUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCAAAUAAACACGUCAUAAACCGUUGACUAUCUUCAAGUGCCCCCC-- --.(((.(((.((((((......((((((.(((((((((((....))))))))..))).)))))).......(((........))))))))).))).....)))....-- ( -30.40, z-score = -1.57, R) >droPer1.super_0 3425748 108 - 11822988 --CGGUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCAAAUAAACCCGUCAUAAACCGUUGACUAUCUUCAAGUGCCCCCCCC --..........((.(((.....((((((.(((((((((((....))))))))..))).)))))).........((((........)))).........)))))...... ( -30.20, z-score = -1.76, R) >droWil1.scaffold_181130 12072052 84 + 16660200 ----------UUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUU-CAGUGGUUAAUAAAUACGUCAUAAGCCGC---CCAACUCCA------------ ----------((((.(..........(((((..((((((((....))))))))...-)))))((........))..........).---)))).....------------ ( -19.80, z-score = -0.93, R) >consensus CGCACUCGAGUUGGUCACAUAAAUGUCACUGCAUGUGGCACAAUUGUGUCGCAAUUGUUGUGGCGAAUAAACACGUCAUAAACCGUUGGCUAUCUUCAA_UGCCUCGU__ .......(((.(((((.......((((((.(((((((((((....))))))))..))).))))))........((........))..))))).))).............. (-23.31 = -23.64 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:12 2011