
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,915,553 – 17,915,701 |
| Length | 148 |
| Max. P | 0.995364 |

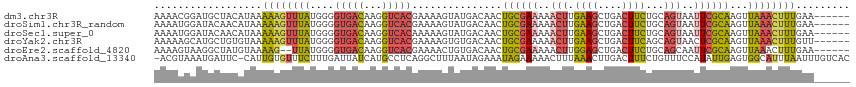
| Location | 17,915,553 – 17,915,663 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.98 |
| Shannon entropy | 0.50819 |
| G+C content | 0.36147 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -13.30 |
| Energy contribution | -13.67 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
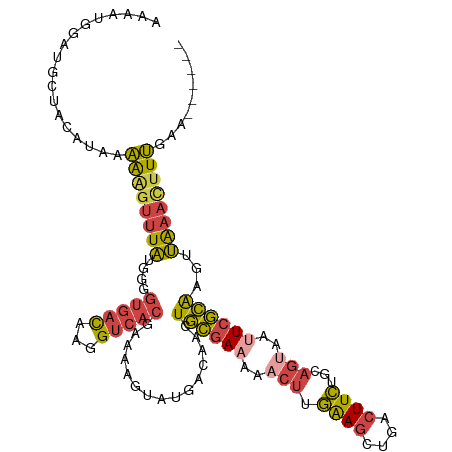
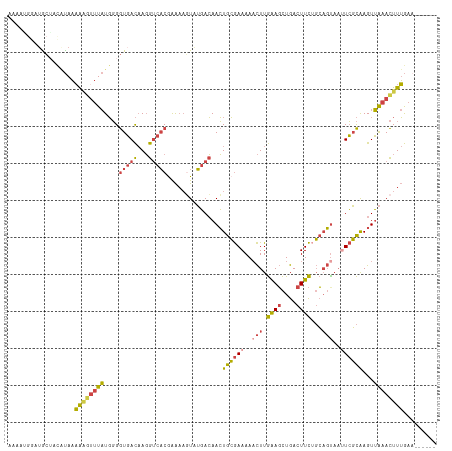
>dm3.chr3R 17915553 110 + 27905053 AAAACGGAUGCUACAUAAAAAGUUUAUGGGGUGACAAGGUCACGAAAAGUAUGACAACUGCGAAAAACUUGAAGCUGACUUCUGCAGUAAUUCGCAAGUUAAACUUUGAA------ .............(((((.....)))))..(((((...)))))..(((((.((((...((((((..(((.((((....))))...)))..)))))).)))).)))))...------ ( -23.70, z-score = -1.21, R) >droSim1.chr3R_random 888262 110 + 1307089 AAAAUGGAUACAACAUAAAAAGUUUAUGGGGUGACAAGGUCACGAAAAGUAUGACAACUGCGAAAAACUUGAAGCUGACUUCUGCAGUAAUUCGCAAGUUAAACUUUGAA------ .............(((((.....)))))..(((((...)))))..(((((.((((...((((((..(((.((((....))))...)))..)))))).)))).)))))...------ ( -23.70, z-score = -1.65, R) >droSec1.super_0 18284346 110 + 21120651 AAAAUGGAUACAACAUAAAAAGUUUAUGGGGUGACAAGGUCACAAAAAGUAUGACAACUGCGAAAAACUUGAAGCUGACUUCUGCAGUAAUUCGCAAGUUAAACUUUGAA------ .............(((((.....)))))..(((((...)))))..(((((.((((...((((((..(((.((((....))))...)))..)))))).)))).)))))...------ ( -23.70, z-score = -1.75, R) >droYak2.chr3R 18755244 110 + 28832112 AAAAAGCAUGCUGUGUAAAAAGUUUAUGGGGUGACAAGGUCACGAAAAGUGUGACAACUGCGAAAAACUUGAAGCUGACUUCAGCAGUAACUCGCAAGUUAAACUUUGUU------ ......(((.(((((.........))))).)))(((((((((((.....))))))((((((((...(((....((((....)))))))...)))).))))....))))).------ ( -26.10, z-score = -0.55, R) >droEre2.scaffold_4820 308382 108 - 10470090 AAAAGUAAGGCUAUGUAAAAG--UUAUGGGGUGACAAGGUCACGAAAACUGUGACAACUGCGAAAAACUUGGAGCUGACUUCUGCAGCAAUUCGCAAGUUAAACUUUGAA------ .(((((.......((((...(--((((((.(((((...))))).....)))))))...))))...((((((((((((.(....)))))...)).))))))..)))))...------ ( -23.80, z-score = -0.43, R) >droAna3.scaffold_13340 8919593 114 + 23697760 -ACGUAAAUGAUUC-CAUUGUGUUUCUUUGAUUAUCAUGCCUCAGGCUUUAAUAGAAAUAGAAAAACUUUAAACUUGACUUUCUGUUUCCAUAUUGAGUGGCAUUUAAUUUGUCAC -......(((((..-((..(.....)..))...)))))(((...))).((((((((((((((((..(.........)..))))))))))..))))))((((((.......)))))) ( -20.10, z-score = -0.76, R) >consensus AAAAUGGAUGCUACAUAAAAAGUUUAUGGGGUGACAAGGUCACGAAAAGUAUGACAACUGCGAAAAACUUGAAGCUGACUUCUGCAGUAAUUCGCAAGUUAAACUUUGAA______ ..................((((((((....(((((...)))))...............((((((..(((.((((....))))...)))..))))))...))))))))......... (-13.30 = -13.67 + 0.37)
| Location | 17,915,553 – 17,915,663 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.98 |
| Shannon entropy | 0.50819 |
| G+C content | 0.36147 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -14.48 |
| Energy contribution | -15.85 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17915553 110 - 27905053 ------UUCAAAGUUUAACUUGCGAAUUACUGCAGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCAUACUUUUCGUGACCUUGUCACCCCAUAAACUUUUUAUGUAGCAUCCGUUUU ------...(((((.(((((.(((((..(((...((((....)))).)))..))))))))))....)))))..(((((...)))))..(((((.....)))))............. ( -24.80, z-score = -3.28, R) >droSim1.chr3R_random 888262 110 - 1307089 ------UUCAAAGUUUAACUUGCGAAUUACUGCAGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCAUACUUUUCGUGACCUUGUCACCCCAUAAACUUUUUAUGUUGUAUCCAUUUU ------...(((((.(((((.(((((..(((...((((....)))).)))..))))))))))....)))))..(((((...)))))..(((((.....)))))............. ( -24.80, z-score = -3.60, R) >droSec1.super_0 18284346 110 - 21120651 ------UUCAAAGUUUAACUUGCGAAUUACUGCAGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCAUACUUUUUGUGACCUUGUCACCCCAUAAACUUUUUAUGUUGUAUCCAUUUU ------...((((((((..(((((((..(((...((((....)))).)))..)))))))..((((((.....))))))............)))))))).................. ( -24.80, z-score = -3.38, R) >droYak2.chr3R 18755244 110 - 28832112 ------AACAAAGUUUAACUUGCGAGUUACUGCUGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCACACUUUUCGUGACCUUGUCACCCCAUAAACUUUUUACACAGCAUGCUUUUU ------...((((((((..(((((((..(((..(((((....))))))))..)))))))..(((((.......)))))............)))))))).................. ( -25.90, z-score = -2.48, R) >droEre2.scaffold_4820 308382 108 + 10470090 ------UUCAAAGUUUAACUUGCGAAUUGCUGCAGAAGUCAGCUCCAAGUUUUUCGCAGUUGUCACAGUUUUCGUGACCUUGUCACCCCAUAA--CUUUUACAUAGCCUUACUUUU ------...((((((.((((.(((((..(((...((.(....)))..)))..)))))))))(((((.......))))).............))--))))................. ( -19.60, z-score = -1.08, R) >droAna3.scaffold_13340 8919593 114 - 23697760 GUGACAAAUUAAAUGCCACUCAAUAUGGAAACAGAAAGUCAAGUUUAAAGUUUUUCUAUUUCUAUUAAAGCCUGAGGCAUGAUAAUCAAAGAAACACAAUG-GAAUCAUUUACGU- (((.........(((((.(.....(((((((.((((((.............)))))).)))))))........).)))))(.....).......)))....-.............- ( -15.54, z-score = 0.47, R) >consensus ______UUCAAAGUUUAACUUGCGAAUUACUGCAGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCAUACUUUUCGUGACCUUGUCACCCCAUAAACUUUUUAUGUAGCAUCCAUUUU .........((((((((..(((((((..(((...((((....)))).)))..)))))))..(((((.......)))))............)))))))).................. (-14.48 = -15.85 + 1.37)
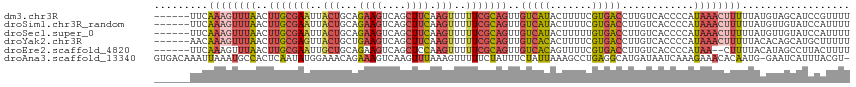
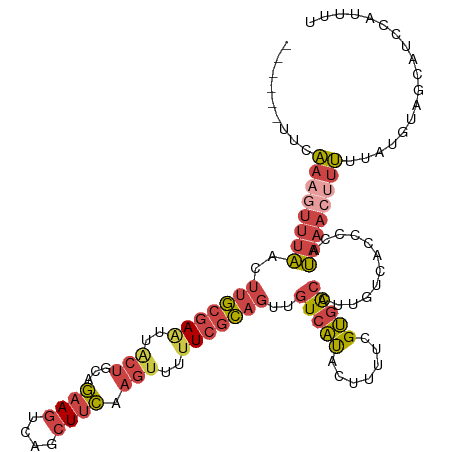
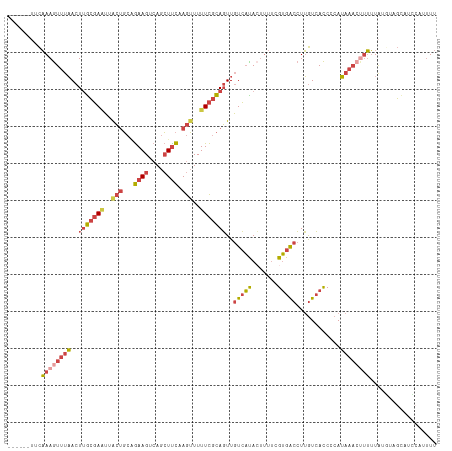
| Location | 17,915,590 – 17,915,701 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 69.01 |
| Shannon entropy | 0.55635 |
| G+C content | 0.38296 |
| Mean single sequence MFE | -21.69 |
| Consensus MFE | -12.81 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17915590 111 - 27905053 ----------GAGCUCUUUUUUCCUUUUUUUUUUGUUCGUGUAUGCCCUUCAAAGUUUAACUUGCGAAUUACUGCAGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCAUACUUUUCGUGACC ----------((((....................)))).............(((((.(((((.(((((..(((...((((....)))).)))..))))))))))....)))))........ ( -21.45, z-score = -1.41, R) >droSim1.chr3R_random 888299 108 - 1307089 ----------GAGCUCUUUUUUCC---AUUUUUUGUUCGUGUAUGCCCUUCAAAGUUUAACUUGCGAAUUACUGCAGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCAUACUUUUCGUGACC ----------((((..........---.......)))).............(((((.(((((.(((((..(((...((((....)))).)))..))))))))))....)))))........ ( -21.63, z-score = -1.44, R) >droSec1.super_0 18284383 108 - 21120651 ----------GAGCUCUUUUUUCC---AUUUUUUGUUCGUGUAUGCCCUUCAAAGUUUAACUUGCGAAUUACUGCAGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCAUACUUUUUGUGACC ----------((((..........---.......))))....................((((.(((((..(((...((((....)))).)))..)))))))))((((((.....)))))). ( -22.83, z-score = -1.81, R) >droYak2.chr3R 18755281 97 - 28832112 ----------GAGCUCUUUUUCC----AUUUUUUUCACA----------ACAAAGUUUAACUUGCGAGUUACUGCUGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCACACUUUUCGUGACC ----------.............----.......((((.----------..(((((.(((((.(((((..(((..(((((....))))))))..))))))))))....)))))..)))).. ( -23.10, z-score = -2.31, R) >droEre2.scaffold_4820 308417 99 + 10470090 ----------GAGCGCUGGUUGC----AUUUUUUGUUU--------CCUUCAAAGUUUAACUUGCGAAUUGCUGCAGAAGUCAGCUCCAAGUUUUUCGCAGUUGUCACAGUUUUCGUGACC ----------((((...((..((----(.....)))..--------))......))))((((.(((((..(((...((.(....)))..)))..)))))))))(((((.......))))). ( -23.10, z-score = -0.26, R) >droAna3.scaffold_13340 8919628 117 - 23697760 AAACUUACUGGAGGCCAUUUUUGAACGUACUAUUAUCUG----UGACAAAUUAAAUGCCACUCAAUAUGGAAACAGAAAGUCAAGUUUAAAGUUUUUCUAUUUCUAUUAAAGCCUGAGGCA ..........((((.(((((.......(((........)----)).......))))).).)))...(((((((.((((((.............)))))).)))))))....(((...))). ( -18.06, z-score = 0.67, R) >consensus __________GAGCUCUUUUUUCC___AUUUUUUGUUCG____UGCCCUUCAAAGUUUAACUUGCGAAUUACUGCAGAAGUCAGCUUCAAGUUUUUCGCAGUUGUCAUACUUUUCGUGACC ..........................................................((((.(((((..(((...((((....)))).)))..)))))))))(((((.......))))). (-12.81 = -13.40 + 0.59)

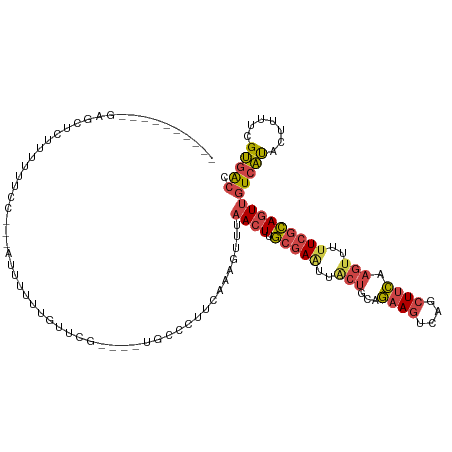
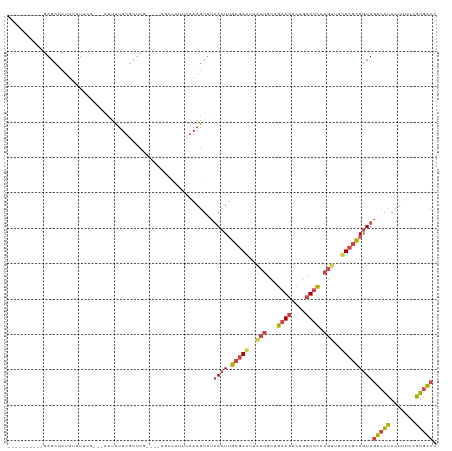
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:08 2011