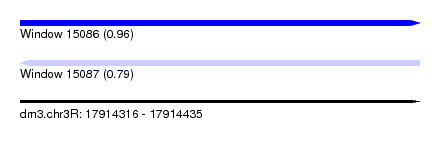
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,914,316 – 17,914,435 |
| Length | 119 |
| Max. P | 0.958730 |

| Location | 17,914,316 – 17,914,435 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.16 |
| Shannon entropy | 0.38237 |
| G+C content | 0.28249 |
| Mean single sequence MFE | -16.60 |
| Consensus MFE | -10.94 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
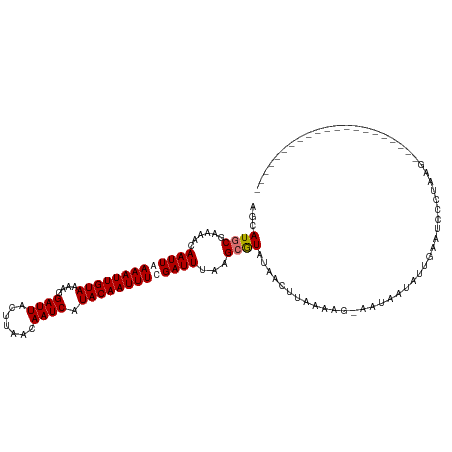
>dm3.chr3R 17914316 119 + 27905053 AGCAUGCGAAAACAAUUAAAAUUGUAAAAGGAUUACUUAACAAUCAUACAAUUUUGAUUUAAGCGUAUAACUUAAGAG-AAUAGUAUUGAUUCCCGGACAUCCAAGAAUAAAAACCUAAG ...(((((.....(((((((((((((....((((.......)))).)))))))))))))....)))))..........-..(((.....((((..((....))..))))......))).. ( -19.50, z-score = -2.43, R) >droEre2.scaffold_4820 307193 99 - 10470090 AACAUGCGAAAACAAUUAAAAUUGUAAAAGGAUUACUUAACAAUCAUACAAUUUCGAUUUAAGAGUAUGACCCAAAAAUAAUAAGAACACAAGCAUAAG--------------------- ..(((((......(((..((((((((....((((.......)))).))))))))..))).....)))))..............................--------------------- ( -13.40, z-score = -2.97, R) >droYak2.chr3R 18754017 98 + 28832112 AGCAUGCGAAAACAAUUAAAAUUGUAAAAGGAUUACUUAGCAAUCAUACAAUUUCGAUUUAAGCAUCGAACUUAGAGG-AAUAAAACUGCAGCCUUAAG--------------------- .((.((((.......(..((((((((....((((.......)))).))))))))..)((((((.......))))))..-........))))))......--------------------- ( -13.60, z-score = -0.33, R) >droSim1.chr3R_random 887091 98 + 1307089 AGCAUGCGAAAACAAUUAAAAUUGUAAAAGGAUUACUUAACAAUCAUACAAUUUUGAUUUAAGCGUAUAACUUAAAGG-UAUAGUGUUGAUUCACUAAG--------------------- ...(((((.....(((((((((((((....((((.......)))).)))))))))))))....)))))..........-..(((((......)))))..--------------------- ( -19.90, z-score = -2.74, R) >consensus AGCAUGCGAAAACAAUUAAAAUUGUAAAAGGAUUACUUAACAAUCAUACAAUUUCGAUUUAAGCGUAUAACUUAAAAG_AAUAAUAUUGAAUCCCUAAG_____________________ ...((((......(((((((((((((....((((.......)))).)))))))))))))...))))...................................................... (-10.94 = -11.50 + 0.56)

| Location | 17,914,316 – 17,914,435 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.16 |
| Shannon entropy | 0.38237 |
| G+C content | 0.28249 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -9.98 |
| Energy contribution | -9.60 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
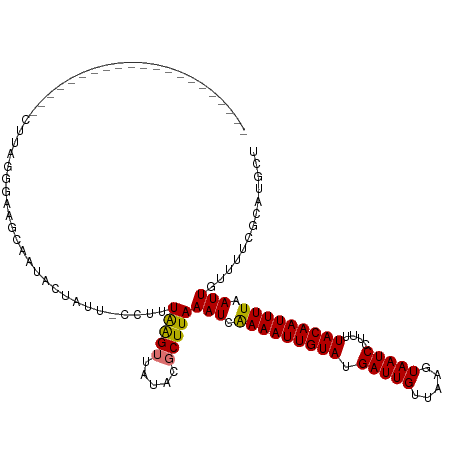
>dm3.chr3R 17914316 119 - 27905053 CUUAGGUUUUUAUUCUUGGAUGUCCGGGAAUCAAUACUAUU-CUCUUAAGUUAUACGCUUAAAUCAAAAUUGUAUGAUUGUUAAGUAAUCCUUUUACAAUUUUAAUUGUUUUCGCAUGCU ....(((.((.((((((((....)))))))).)).)))...-...((((((.....))))))...(((((((((.(((((.....)))))....)))))))))................. ( -21.00, z-score = -1.63, R) >droEre2.scaffold_4820 307193 99 + 10470090 ---------------------CUUAUGCUUGUGUUCUUAUUAUUUUUGGGUCAUACUCUUAAAUCGAAAUUGUAUGAUUGUUAAGUAAUCCUUUUACAAUUUUAAUUGUUUUCGCAUGUU ---------------------..(((((.((((..((((.......)))).))))......(((..((((((((.(((((.....)))))....))))))))..)))......))))).. ( -16.40, z-score = -2.19, R) >droYak2.chr3R 18754017 98 - 28832112 ---------------------CUUAAGGCUGCAGUUUUAUU-CCUCUAAGUUCGAUGCUUAAAUCGAAAUUGUAUGAUUGCUAAGUAAUCCUUUUACAAUUUUAAUUGUUUUCGCAUGCU ---------------------.....((((((.........-....(((((.....)))))(((..((((((((.((((((...))))))....))))))))..)))......))).))) ( -20.20, z-score = -2.12, R) >droSim1.chr3R_random 887091 98 - 1307089 ---------------------CUUAGUGAAUCAACACUAUA-CCUUUAAGUUAUACGCUUAAAUCAAAAUUGUAUGAUUGUUAAGUAAUCCUUUUACAAUUUUAAUUGUUUUCGCAUGCU ---------------------....(((((..((((.....-..(((((((.....)))))))..(((((((((.(((((.....)))))....)))))))))...)))))))))..... ( -17.10, z-score = -2.32, R) >consensus _____________________CUUAGGGAAGCAAUACUAUU_CCUUUAAGUUAUACGCUUAAAUCAAAAUUGUAUGAUUGUUAAGUAAUCCUUUUACAAUUUUAAUUGUUUUCGCAUGCU ..............................................(((((.....)))))(((.(((((((((.(((((.....)))))....))))))))).)))............. ( -9.98 = -9.60 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:06 2011