| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,894,066 – 17,894,181 |
| Length | 115 |
| Max. P | 0.925051 |

| Location | 17,894,066 – 17,894,181 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 135 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Shannon entropy | 0.48636 |
| G+C content | 0.45429 |
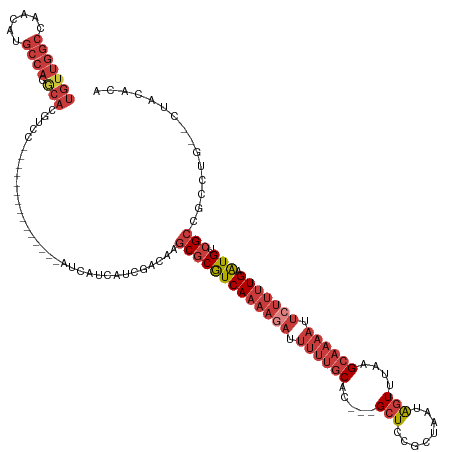
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -17.27 |
| Energy contribution | -18.99 |
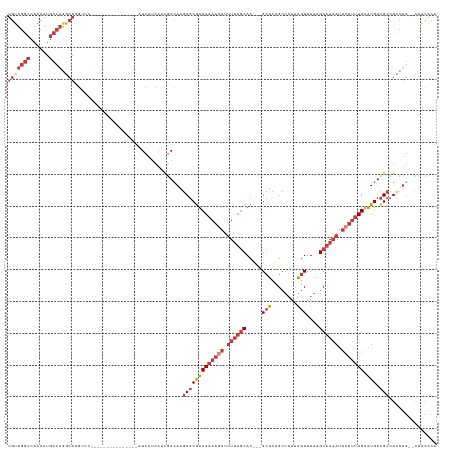
| Covariance contribution | 1.71 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
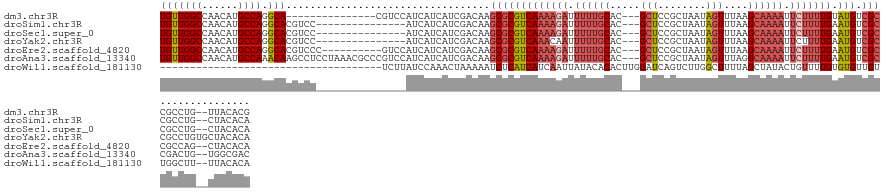
>dm3.chr3R 17894066 115 + 27905053 UGUUGGCCAACAUGCCAGGCA---------------CGUCCAUCAUCAUCGACAAGCGCGUCAAAAGAUUUUUGCAC---GCUCCGCUAAUAGUUUAAGCAAAAUUCUUUUGUAUGUCGCCGCCUG--UUACACG ....(((......)))((((.---------------.(((.((....)).)))..(((((((((((((.((((((..---(((........)))....)))))).))))))).))).))).)))).--....... ( -31.00, z-score = -2.02, R) >droSim1.chr3R 17719559 115 + 27517382 UGUUGGCCAACAUGCCAGGCACGUCC---------------AUCAUCAUCGACAAGCGCGUCAAAAGAUUUUUGCAC---GCUCCGCUAAUAGUUUAAGCAAAAUUCUUUUGAAUGUCGCCGCCUG--CUACACA ....(((......)))((((..(((.---------------((....)).)))..(((((((((((((.((((((..---(((........)))....)))))).))))))).))).))).)))).--....... ( -31.00, z-score = -1.72, R) >droSec1.super_0 18263415 115 + 21120651 UGUUGGCCAACAUGCCAGGCACGUCC---------------AUCAUCAUCGACAAGCGCGUCAAAAGAUUUUUGCAC---GCUCCGCUAAUAGUUUAAGCAAAAUUCUUUUGAAUGUCGCCGCCUG--CUACACA ....(((......)))((((..(((.---------------((....)).)))..(((((((((((((.((((((..---(((........)))....)))))).))))))).))).))).)))).--....... ( -31.00, z-score = -1.72, R) >droYak2.chr3R 18732547 117 + 28832112 UGUUGGCCAACAUGCCAGGCACGUCC---------------AUCAUCAUCGACAAGCGCGUCAAACAAUUUUUGCAC---GCUCCGCUAAUAGUUUAAGCAAAAUUCUUUUGAAUGUCGCCGCCUGUGCUACACA ((((....)))).(((((((..(((.---------------((....)).)))..((((((((((....((((((..---(((........)))....))))))....)))).))).))).))))).))...... ( -26.60, z-score = -0.13, R) >droEre2.scaffold_4820 285320 120 - 10470090 UGUUGGCCAACAUGCCAGGCACGUCCC----------GUCCAUCAUCAUCGACAAGCGCGUCAAAAGAUUUUUGCAC---GCUCCGCUAAUAGUUUAAGCAAAAUUCUUUUGAAUGUCGCCGCCAG--CUACACA .((((((..........((.(((...)----------))))..............(((((((((((((.((((((..---(((........)))....)))))).))))))).))).))).)))))--)...... ( -31.70, z-score = -1.91, R) >droAna3.scaffold_13340 8897087 130 + 23697760 UGUUGGCCAACAUGCCAAACAAGCCUCCUAAACGCCCGUCCAUCAUCAUCGACAAGCGCGUCAAAAGAUUUUUGCAC---GCUCCGCUAAUAGUUUAGGCAAAAUUCUUUUGAAUGUCGCCGACUG--UGGCGAC (((((((......))).))))...........(((..(((.((....)).)))..)))..((((((((.((((((..---(((........)))....)))))).))))))))..(((((((....--))))))) ( -35.60, z-score = -1.83, R) >droWil1.scaffold_181130 12005037 96 + 16660200 -------------------------------------UCUUAUCCAAACUAAAAAUCUCAUCAUCAAUUAUACACACUUGGAUCAGUCUUGGCUUUUAGCUAUACUGUUUUGUGUGUUGUUGGCUU--UUACACA -------------------------------------..........................(((((...((((((..(((.((((..((((.....)))).))))))).)))))).)))))...--....... ( -18.70, z-score = -2.06, R) >consensus UGUUGGCCAACAUGCCAGGCACGUCC_______________AUCAUCAUCGACAAGCGCGUCAAAAGAUUUUUGCAC___GCUCCGCUAAUAGUUUAAGCAAAAUUCUUUUGAAUGUCGCCGCCUG__CUACACA ..(((((......))))).....................................(((((((((((((.((((((.....(((........)))....)))))).))))))).))).)))............... (-17.27 = -18.99 + 1.71)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:03 2011