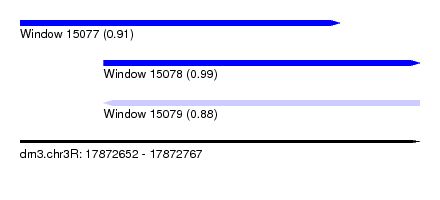
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,872,652 – 17,872,767 |
| Length | 115 |
| Max. P | 0.992516 |

| Location | 17,872,652 – 17,872,744 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 89.12 |
| Shannon entropy | 0.17211 |
| G+C content | 0.42033 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
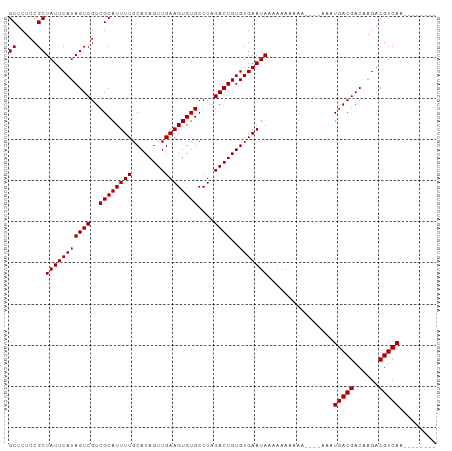
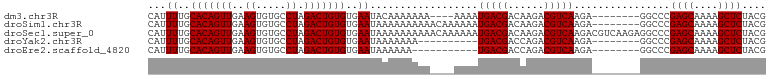
>dm3.chr3R 17872652 92 + 27905053 GCUCUCCGCUAUUCAUAGUCGUCGCAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUACAAAAAAAA----AAAUGACGACAAGACGUCAA-------- ((.....))(((((((((((..((((((((........))))))))....))))..))))))).........----...(((((......))))).-------- ( -21.40, z-score = -1.15, R) >droSim1.chr3R 17696724 96 + 27517382 GCUCUCCGCUAUUCAUAGUCGUCGCAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAAAAAACAAAAAAUGACGACAAGACGUCAA-------- ((.....))(((((((((((..((((((((........))))))))....))))..)))))))................(((((......))))).-------- ( -21.70, z-score = -1.46, R) >droSec1.super_0 18242206 104 + 21120651 GCUCUCCGCUAUUCAUAGUCGUCGCAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAAAAAACAAAAAAUGACGACAAGACGUCAAGACGUCAA ((.....))........(((((((...((..(((((((..((.....)).)))))))..))..................)))))))..(((((....))))).. ( -26.80, z-score = -2.21, R) >droYak2.chr3R 18710473 86 + 28832112 GCUCUCCGCUAUUCAUAGUCGUCGCAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAAA----------UGACGACCAGACGUCAA-------- ((.....))(((((((((((..((((((((........))))))))....))))..)))))))......----------(((((......))))).-------- ( -21.70, z-score = -1.37, R) >droEre2.scaffold_4820 263696 85 - 10470090 GCUCUCCGCUAUUCAUAGUCGUCGCAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAA-----------UGACGACCAGACGUCAA-------- ((.....))(((((((((((..((((((((........))))))))....))))..))))))).....-----------(((((......))))).-------- ( -21.70, z-score = -1.34, R) >consensus GCUCUCCGCUAUUCAUAGUCGUCGCAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAAAAAA____AAAUGACGACAAGACGUCAA________ ((.....))(((((((((((..((((((((........))))))))....))))..)))))))................(((((......)))))......... (-21.52 = -21.52 + -0.00)

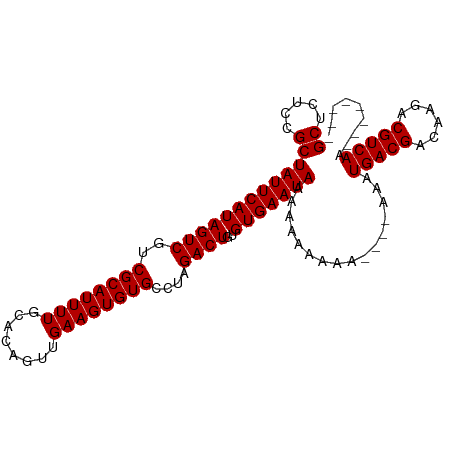
| Location | 17,872,676 – 17,872,767 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Shannon entropy | 0.17378 |
| G+C content | 0.43594 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17872676 91 + 27905053 CAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUACAAAAAAA----AAAAUGACGACAAGACGUCAAGA--------GGCCCGAGCAAAAGCUCUACG ...((..(((((((..((.....)).)))))))..))..........----....(((((......)))))...--------.....((((....)))).... ( -22.80, z-score = -2.36, R) >droSim1.chr3R 17696748 95 + 27517382 CAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAAAAAACAAAAAAUGACGACAAGACGUCAAGA--------GGCCCGAGCAAAAGCUCUACG ...((..(((((((..((.....)).)))))))..))..................(((((......)))))...--------.....((((....)))).... ( -22.80, z-score = -2.51, R) >droSec1.super_0 18242230 103 + 21120651 CAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAAAAAACAAAAAAUGACGACAAGACGUCAAGACGUCAAGAGGCCCGAGCAAAAGCUCUACG ...((..(((((((..((.....)).)))))))..))...........................(((((....))))).........((((....)))).... ( -24.10, z-score = -2.03, R) >droYak2.chr3R 18710497 85 + 28832112 CAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAAA----------UGACGACCAGACGUCAAGA--------GGCCCGAGCAAAAGCUCUACG ...((..(((((((..((.....)).)))))))..))........----------(((((......)))))...--------.....((((....)))).... ( -22.80, z-score = -2.44, R) >droEre2.scaffold_4820 263720 84 - 10470090 CAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAA-----------UGACGACCAGACGUCAAGA--------GGCCCGAGCAAAAGCUCUACG ...((..(((((((..((.....)).)))))))..)).......-----------(((((......)))))...--------.....((((....)))).... ( -22.80, z-score = -2.41, R) >consensus CAUUUUGCACAGUUGAAGUGUGCCUAGACUGUGUGAAUAAAAAAAAA____AAAAUGACGACAAGACGUCAAGA________GGCCCGAGCAAAAGCUCUACG ...((..(((((((..((.....)).)))))))..))..................(((((......)))))................((((....)))).... (-22.68 = -22.68 + -0.00)
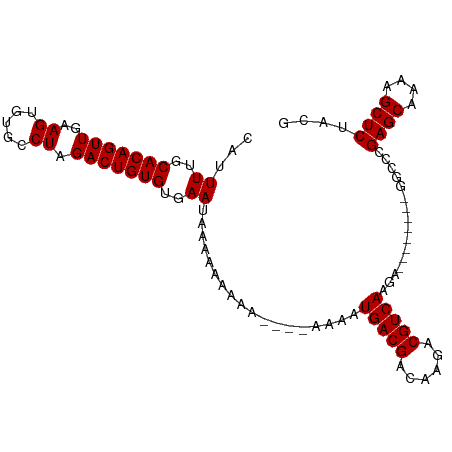
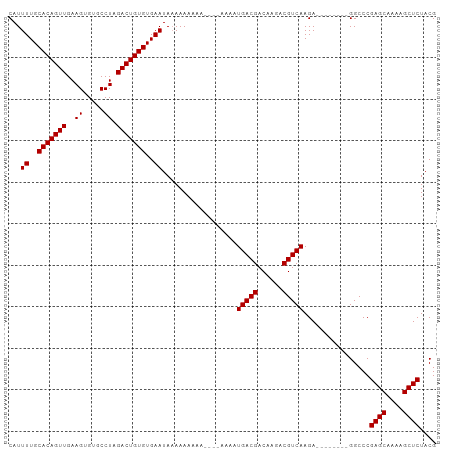
| Location | 17,872,676 – 17,872,767 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Shannon entropy | 0.17378 |
| G+C content | 0.43594 |
| Mean single sequence MFE | -21.04 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17872676 91 - 27905053 CGUAGAGCUUUUGCUCGGGCC--------UCUUGACGUCUUGUCGUCAUUUU----UUUUUUUGUAUUCACACAGUCUAGGCACACUUCAACUGUGCAAAAUG ....((((....))))((((.--------...(((((......)))))....----......(((......)))))))..(((((.......)))))...... ( -20.20, z-score = -1.29, R) >droSim1.chr3R 17696748 95 - 27517382 CGUAGAGCUUUUGCUCGGGCC--------UCUUGACGUCUUGUCGUCAUUUUUUGUUUUUUUUUUAUUCACACAGUCUAGGCACACUUCAACUGUGCAAAAUG .((.((((....))))..)).--------...(((((......)))))......................((((((..(((....)))..))))))....... ( -20.00, z-score = -1.38, R) >droSec1.super_0 18242230 103 - 21120651 CGUAGAGCUUUUGCUCGGGCCUCUUGACGUCUUGACGUCUUGUCGUCAUUUUUUGUUUUUUUUUUAUUCACACAGUCUAGGCACACUUCAACUGUGCAAAAUG ....((((....)))).(((..(..(((((....)))))..)..))).......................((((((..(((....)))..))))))....... ( -25.00, z-score = -2.15, R) >droYak2.chr3R 18710497 85 - 28832112 CGUAGAGCUUUUGCUCGGGCC--------UCUUGACGUCUGGUCGUCA----------UUUUUUUAUUCACACAGUCUAGGCACACUUCAACUGUGCAAAAUG .((.((((....))))..)).--------...(((((......)))))----------............((((((..(((....)))..))))))....... ( -20.00, z-score = -1.22, R) >droEre2.scaffold_4820 263720 84 + 10470090 CGUAGAGCUUUUGCUCGGGCC--------UCUUGACGUCUGGUCGUCA-----------UUUUUUAUUCACACAGUCUAGGCACACUUCAACUGUGCAAAAUG .((.((((....))))..)).--------...(((((......)))))-----------...........((((((..(((....)))..))))))....... ( -20.00, z-score = -1.20, R) >consensus CGUAGAGCUUUUGCUCGGGCC________UCUUGACGUCUUGUCGUCAUUUU____UUUUUUUUUAUUCACACAGUCUAGGCACACUUCAACUGUGCAAAAUG .((.((((....))))..))............(((((......)))))......................((((((..(((....)))..))))))....... (-19.76 = -19.76 + -0.00)
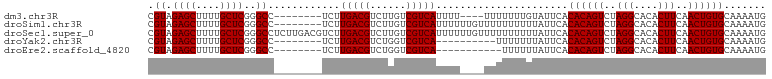
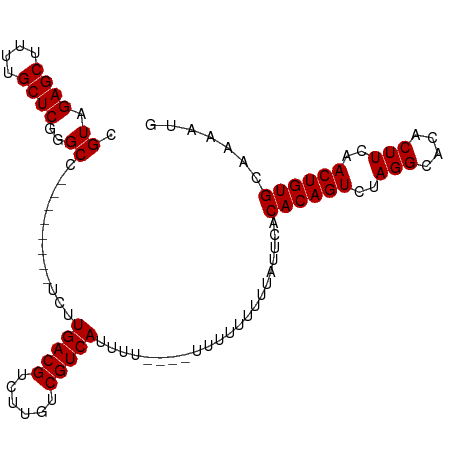
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:59 2011