| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,838,568 – 17,838,663 |
| Length | 95 |
| Max. P | 0.999832 |

| Location | 17,838,568 – 17,838,663 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 72.60 |
| Shannon entropy | 0.57745 |
| G+C content | 0.41629 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.03 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.52 |
| SVM RNA-class probability | 0.999832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
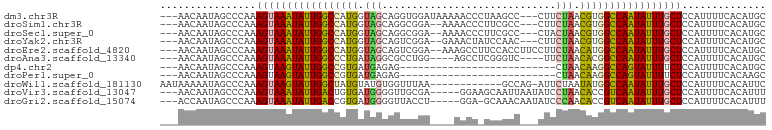
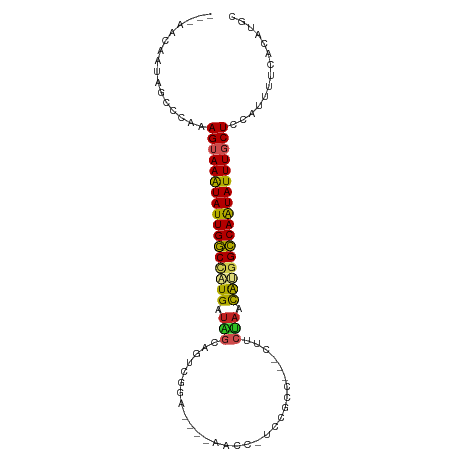
>dm3.chr3R 17838568 95 + 27905053 ---AACAAUAGCCCAAAGUAAAUAUUGGCCAUGGUAGCAGGUGGAUAAAAACCCUAAGCC---CUUCUAACGUGGCCAAUAUUUGCUCCAUUUUCACAUGC ---.............(((((((((((((((((.(((.(((.((........)).....)---)).))).))))))))))))))))).............. ( -31.20, z-score = -3.85, R) >droSim1.chr3R 17661133 93 + 27517382 ---AACAAUAGCCCAAAGUAAAUAUUGGCCAUGGUAGCAGGCGGA--AAAACCCUUCGCC---CUUCUAACGUGGCCAAUAUUUGCUCCAUUUUCACAUGC ---.............(((((((((((((((((.(((..((((((--.......))))))---...))).))))))))))))))))).............. ( -35.20, z-score = -5.36, R) >droSec1.super_0 18208922 93 + 21120651 ---AACAAUAGCCCAAAGUAAAUAUUGGCCAUGGUAGCAGGCGGA--AAAACCCUUCGCC---CUACUAACGUGGCCAAUAUUUGCUCCAUUUUCACAUGC ---.............(((((((((((((((((.(((..((((((--.......))))))---...))).))))))))))))))))).............. ( -35.20, z-score = -5.32, R) >droYak2.chr3R 18672400 93 + 28832112 ---AACAAUAGCCCAAAGUAAAUAUUGGCCAUGGUAGCAGUCGGA--GAAACUAUCCAAC---CUUCUAACGUGGCCAAUAUUUGCUCCAUUUUCACAUGC ---.............(((((((((((((((((.(((..((.(((--.......))).))---...))).))))))))))))))))).............. ( -30.70, z-score = -4.98, R) >droEre2.scaffold_4820 227351 96 - 10470090 ---AACAAUAGCCCAAAGUAAAUAUUGGCCAUGGUAGCAGUCGGA--AAAGCCUUCCACCUUCCUUCUAACAUGGCCAAUAUUUGCUCCAUUUUCACAUGC ---.............(((((((((((((((((.(((.((..(((--(.....))))..)).....))).))))))))))))))))).............. ( -29.80, z-score = -4.57, R) >droAna3.scaffold_13340 8843268 90 + 23697760 ---AACAAUAGCCCAAAGUAAAUAUUGGCCCUGAUAGGCGCCUGG----AGCCUCGGGUC----UUCUAACACGGCCAAUAUUUGCUCCAUUUUCACAUGC ---.............((((((((((((((.((.((((.((((((----....)))))).----.)))).)).)))))))))))))).............. ( -30.60, z-score = -3.35, R) >dp4.chr2 6702531 72 + 30794189 ---AACAAUAGCCCAAAGUAAGUAUUGGCCGUGAUGAGAG--------------------------CUAACAAGGCCAGUAUUUUCUCCAUUUUCACAUGC ---.............((.(((((((((((.((.((....--------------------------.)).)).))))))))))).)).............. ( -15.00, z-score = -1.32, R) >droPer1.super_0 527921 72 + 11822988 ---AACAAUAGCCCAAAGUAAGUAUUGGCCGUGAUGAGAG--------------------------CUAACAAGGCCAGUAUUUUCUCCAUUUUCACAAGC ---.............((.(((((((((((.((.((....--------------------------.)).)).))))))))))).)).............. ( -15.00, z-score = -1.57, R) >droWil1.scaffold_181130 11938102 88 + 16660200 AAUAAAAAUAGCCCAAAGUAAGUAUUGGCUAUGUAUGUGGUUUAA------------GCCAG-AUUCUAAUAUGGCCAAUAUUUGCUCCAUUUUCACAUUC ................((((((((((((((((((((.((((....------------)))).-))....)))))))))))))))))).............. ( -26.90, z-score = -4.62, R) >droVir3.scaffold_13047 10573546 93 - 19223366 ---AACAAUAGCCCAAAGUAAAUAUUGACUGUGAUGGGGUUGCGA-----GGAAGCAAUUAAUAUCCUAACACCGUCAAUAUUUGCUCCAUUUUCACAUUU ---.............(((((((((((((.(((.(((((((((..-----....)))......)))))).))).))))))))))))).............. ( -27.40, z-score = -4.01, R) >droGri2.scaffold_15074 973153 92 + 7742996 ---ACCAAUAGCCCAAAGUAAAUAUUGACCGUGAUGGGGUUACCU-----GGA-GCAAACAAUAUCCCAACACCGUCAAUAUUUGCUCCAUUUUCACAUUU ---.............(((((((((((((.(((.((((((....(-----(..-.....))..)))))).))).))))))))))))).............. ( -27.70, z-score = -4.49, R) >consensus ___AACAAUAGCCCAAAGUAAAUAUUGGCCAUGAUAGCAGUCGGA____AACC_UCCGCC___CUUCUAACAUGGCCAAUAUUUGCUCCAUUUUCACAUGC ................(((((((((((((((((.(((.............................))).))))))))))))))))).............. (-21.52 = -21.03 + -0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:54 2011