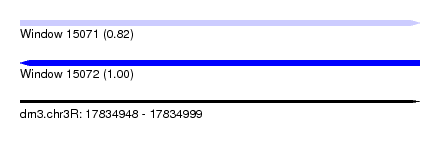
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,834,948 – 17,834,999 |
| Length | 51 |
| Max. P | 0.997764 |

| Location | 17,834,948 – 17,834,999 |
|---|---|
| Length | 51 |
| Sequences | 6 |
| Columns | 55 |
| Reading direction | forward |
| Mean pairwise identity | 74.52 |
| Shannon entropy | 0.47523 |
| G+C content | 0.56744 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -8.08 |
| Energy contribution | -8.97 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
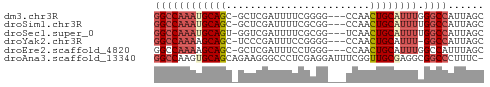
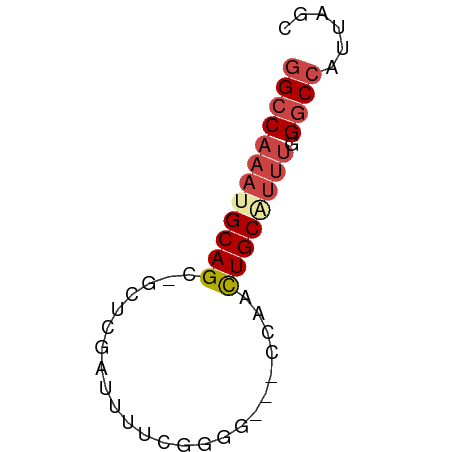
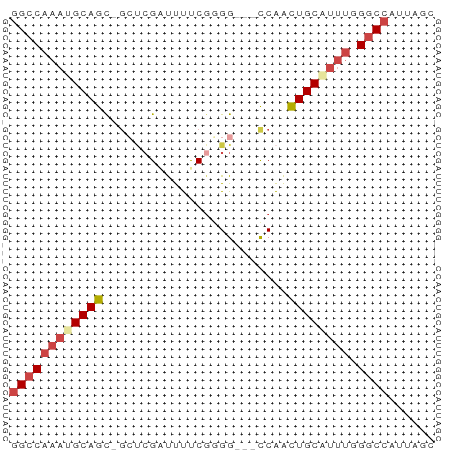
>dm3.chr3R 17834948 51 + 27905053 GCUAAUGGCCCAAAUGCAGUUGG---CCCCGAAAAUCGAGC-GCUGCAUUUGGCC ......((((.(((((((((..(---(..((.....)).))-))))))))))))) ( -20.70, z-score = -2.72, R) >droSim1.chr3R 17657550 51 + 27517382 GCUAAUGGCCAAAAUGCAGUUGG---CCGCGAAAAUCGAGC-GCUGCAUUUGGCC ......(((((((.((((((..(---(..((.....)).))-))))))))))))) ( -20.80, z-score = -2.42, R) >droSec1.super_0 18205294 51 + 21120651 GCUAAUGGCCAAAAUGCAGUUGA---CCGCGAAAAUCGACC-ACUGCAUUUGGCC ......(((((((.((((((.(.---...((.....)).).-))))))))))))) ( -18.70, z-score = -3.79, R) >droYak2.chr3R 18668669 50 + 28832112 GCUAAUGGCC-AAAUGCAGUUGG---CCCCGGAAAUCGGGA-GCUGCUUUUGGCC ......((((-(((.((((((..---.((((.....)))))-))))).))))))) ( -24.40, z-score = -3.08, R) >droEre2.scaffold_4820 223704 51 - 10470090 GCUAAAUGGCCAAAUGCAGUUGG---CCCAGGAAAUCGAGC-GCUGCUUUUGGCC .......(((((((.(((((..(---(.(........).))-))))).))))))) ( -18.60, z-score = -1.41, R) >droAna3.scaffold_13340 8839412 54 + 23697760 -GAAAGGGCCGCCUCGCAACCGAAAUCCUCGAGGGCCCUUCUGCUGCACUUGGCC -..(((((((((...))...(((.....)))..)))))))..(((......))). ( -15.70, z-score = 0.21, R) >consensus GCUAAUGGCCAAAAUGCAGUUGG___CCCCGAAAAUCGAGC_GCUGCAUUUGGCC ......((((.(((.(((((.(..................).))))).))))))) ( -8.08 = -8.97 + 0.89)

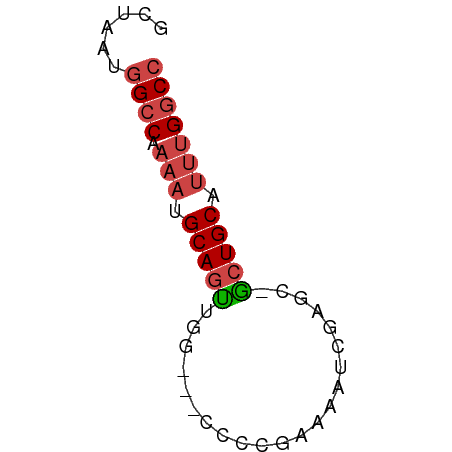
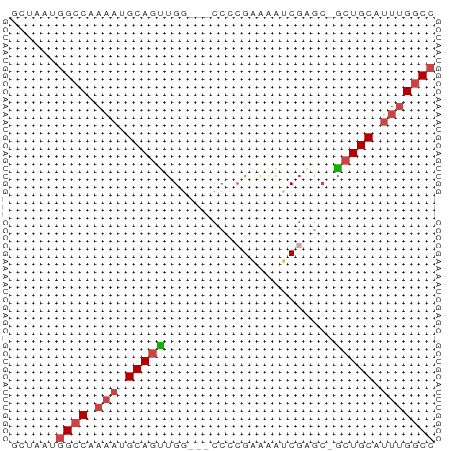
| Location | 17,834,948 – 17,834,999 |
|---|---|
| Length | 51 |
| Sequences | 6 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 74.52 |
| Shannon entropy | 0.47523 |
| G+C content | 0.56744 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -11.06 |
| Energy contribution | -12.01 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.997764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17834948 51 - 27905053 GGCCAAAUGCAGC-GCUCGAUUUUCGGGG---CCAACUGCAUUUGGGCCAUUAGC ((((((((((((.-((((........)))---)...)))))))).))))...... ( -22.90, z-score = -3.12, R) >droSim1.chr3R 17657550 51 - 27517382 GGCCAAAUGCAGC-GCUCGAUUUUCGCGG---CCAACUGCAUUUUGGCCAUUAGC ((((((((((((.-(((((.....)).))---)...))))).)))))))...... ( -22.00, z-score = -3.35, R) >droSec1.super_0 18205294 51 - 21120651 GGCCAAAUGCAGU-GGUCGAUUUUCGCGG---UCAACUGCAUUUUGGCCAUUAGC (((((((((((((-(..((.......)).---.).)))))).)))))))...... ( -25.00, z-score = -5.30, R) >droYak2.chr3R 18668669 50 - 28832112 GGCCAAAAGCAGC-UCCCGAUUUCCGGGG---CCAACUGCAUUU-GGCCAUUAGC (((((((.((((.-(((((.....)))))---....)))).)))-))))...... ( -24.50, z-score = -3.79, R) >droEre2.scaffold_4820 223704 51 + 10470090 GGCCAAAAGCAGC-GCUCGAUUUCCUGGG---CCAACUGCAUUUGGCCAUUUAGC (((((((.((((.-(((((......))))---)...)))).)))))))....... ( -22.90, z-score = -3.44, R) >droAna3.scaffold_13340 8839412 54 - 23697760 GGCCAAGUGCAGCAGAAGGGCCCUCGAGGAUUUCGGUUGCGAGGCGGCCCUUUC- ((((...((((((.((((..((.....)).)))).))))))....)))).....- ( -19.50, z-score = -0.16, R) >consensus GGCCAAAUGCAGC_GCUCGAUUUUCGGGG___CCAACUGCAUUUGGGCCAUUAGC ((((((((((((........................)))))))).))))...... (-11.06 = -12.01 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:53 2011