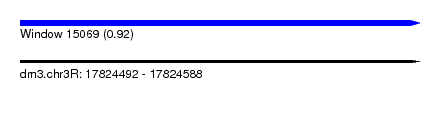
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,824,492 – 17,824,588 |
| Length | 96 |
| Max. P | 0.918618 |

| Location | 17,824,492 – 17,824,588 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.54 |
| Shannon entropy | 0.36144 |
| G+C content | 0.50417 |
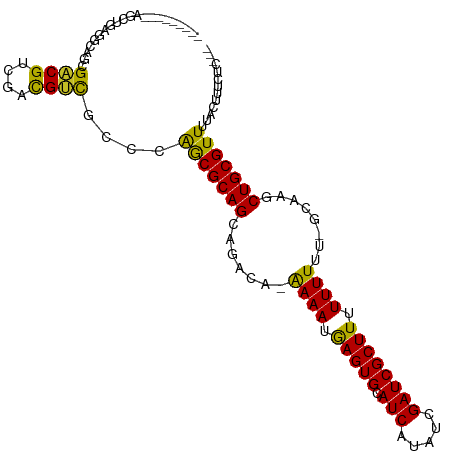
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -20.15 |
| Energy contribution | -19.31 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
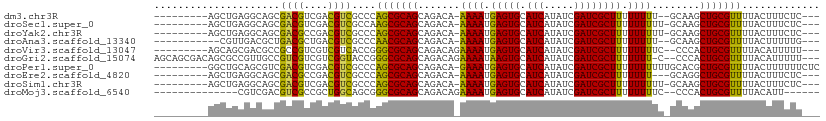
>dm3.chr3R 17824492 96 + 27905053 ---------AGCUGAGGCAGCGACGUCGACGUCGCCCAGCGCAGCAGACA-AAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUUU--GCAAGCUGCGUUUUACUUUCUC--- ---------.((((.(((...((((....)))))))))))(((((...((-((((.(((((.(((.....)))))))).)))))--)...))))).............--- ( -36.10, z-score = -3.03, R) >droSec1.super_0 18194887 97 + 21120651 ---------AGCUGAGGCAGCGACGUCGACGUCGCCAAGCGCAGCAGACA-AAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUUUU-GCAAGCUGCGUUUUACUUUCUC--- ---------....((((..((((((....)))))).(((((((((...((-((((.(((((.(((.....))))))))..)))))-)...))))))))).....))))--- ( -35.30, z-score = -2.65, R) >droYak2.chr3R 18657857 97 + 28832112 ---------AGCUGAGGCAGCGACGCCGACGUCGCCCAGCGCAGCAGACA-AAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUUUU-GCAAGCUGCGUUUUACUUUCUC--- ---------.((((.(((...((((....)))))))))))(((((...((-((((.(((((.(((.....))))))))..)))))-)...))))).............--- ( -34.30, z-score = -2.44, R) >droAna3.scaffold_13340 8829654 94 + 23697760 -----------CGUUGACGCUGACGCUGACGUCGCCCAACGCAGCAGACA-AAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUUU--GCAAGCUGCGUUUUACUUUUUG--- -----------.((((..((.((((....)))))).))))(((((...((-((((.(((((.(((.....)))))))).)))))--)...))))).............--- ( -29.80, z-score = -2.32, R) >droVir3.scaffold_13047 10558816 97 - 19223366 ---------AGCAGCGACGCCGCCGUCGUCGUCACCGGGCGCAGCAGACAGAAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUUUC--CCCACUGCGUUUUACAUUUUU--- ---------.((.((((((....)))))).(((....))))).((((...(((((.(((((.(((.....))))))))..)))))--....)))).............--- ( -25.90, z-score = -0.95, R) >droGri2.scaffold_15074 958489 105 + 7742996 AGCAGCGACAGCGCCGUUGCCGUCGUCGUCGGUACCGGGCGCAGCAGACAGAAAAUAAGUGCAUCAUAUCGAUCGCUUUUUUU-C--CCCACUGCGUUUUACAUUUUU--- .((((.(...((((((.(((((.......))))).).)))))........(((((.(((((.(((.....)))))))).))))-)--..).)))).............--- ( -32.90, z-score = -2.11, R) >droPer1.super_0 511572 101 + 11822988 ---------GGCUGCAGCGUCGACGUCGACGUCGCCCAGCGCAGCAGACA-GAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUUUUUGCACGCUGCGUUUUACUUUUUUCUC ---------((..((.((((((....)))))).))))((((((((...((-((((.(((((.(((.....))))))))...))))))...))))))))............. ( -35.70, z-score = -2.74, R) >droEre2.scaffold_4820 212813 95 - 10470090 ---------AGCUGAGGCAGCGACGCCGACGUCGCCCAGCGCAGCAGACA-AAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUU---GCAGGCUGCGUUUUACUUUCUC--- ---------.((((.(((...((((....)))))))))))(((((...((-((((.(((((.(((.....)))))))))))))---)...))))).............--- ( -34.90, z-score = -2.31, R) >droSim1.chr3R 17645124 97 + 27517382 ---------AGCUGAGGCAGCGACGUCGACGUCGCCCAGCGCAGCAGACA-AAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUUUU-GCAAGCUGCGUUUUACUUUCUC--- ---------.((((.(((...((((....)))))))))))(((((...((-((((.(((((.(((.....))))))))..)))))-)...))))).............--- ( -35.10, z-score = -2.71, R) >droMoj3.scaffold_6540 32036820 89 + 34148556 --------------CGUCGACGUCGCCGCUGGCAGCGGGCGCAGCAGACAGAAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUUUC--CCCACUGCGUUUUACAUU------ --------------.((.((((.((((((.....)).))))).((((...(((((.(((((.(((.....))))))))..)))))--....)))))))..))...------ ( -24.70, z-score = -0.19, R) >consensus _________AGCUGAGGCAGCGACGUCGACGUCGCCCAGCGCAGCAGACA_AAAAUGAGUGCAUCAUAUCGAUCGCUUUUUUUUU_GCAAGCUGCGUUUUACUUUCUC___ .....................((((....))))....(((((((.......((((.(((((.(((.....)))))))).))))........)))))))............. (-20.15 = -19.31 + -0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:51 2011