
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,820,621 – 17,820,718 |
| Length | 97 |
| Max. P | 0.740063 |

| Location | 17,820,621 – 17,820,718 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.55 |
| Shannon entropy | 0.53086 |
| G+C content | 0.46002 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -13.97 |
| Energy contribution | -14.68 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17820621 97 + 27905053 ---------CAUAUUCUCGCACUCC-GUGGCCAAAUGCAUUUUGCAUUAAAUUGCAUAACUUUUGUGCGGCAAUUAGAGUU---CUCUGG-CUGGCCAAGACGGUUGGGAA ---------....((((((.(((..-.(((((((((((.....))))).((((((...((....))...))))))(((...---.)))..-.))))))....))))))))) ( -28.20, z-score = -1.11, R) >droGri2.scaffold_15074 954487 77 + 7742996 -----------------UUGCCUUGUGCGGCC-UAUGCAUUUUACAUUAAAUUGCAUAACUUUUGU--GGCAAUUAGACGCG----CCAU--UCGCAAGGAAA-------- -----------------...(((((((.(((.-((((((.((((...)))).))))))......((--(.........))))----))..--.)))))))...-------- ( -21.80, z-score = -1.53, R) >droPer1.super_0 508233 85 + 11822988 ------------------GCGGGCGAGCGACCACCUGCAUUUUGCAUUAAAUUGCAUAACUUUUGU--GGCAAUUAGACGU-----UCGGCCCGGCCAAGACGA-AGGAUA ------------------.((((((((((......(((.....)))...((((((...((....))--.))))))...)))-----)).))))).((.......-.))... ( -26.10, z-score = -1.36, R) >dp4.chr2 6682829 85 + 30794189 ------------------GCGGGCGAGCGACCACCUGCAUUUUGCAUUAAAUUGCAUAACUUUUGU--GGCAAUUAGACGU-----UCGGCCCGGCCAAGACGA-AGGAUA ------------------.((((((((((......(((.....)))...((((((...((....))--.))))))...)))-----)).))))).((.......-.))... ( -26.10, z-score = -1.36, R) >droAna3.scaffold_13340 8825992 107 + 23697760 UUUCUCCACCGUAUUUCAAGUCUCC--UGGCC--GUGCAUUUUGCAUUAAAUUGCACAACUUUUGUGUGGCAAUUAGACCUUCUGUGUGUUCUGGCCAAGACGCCGGGAUA ...................((((..--..(((--(..((...((((......)))).......))..))))....))))........(((((((((......))))))))) ( -28.20, z-score = -1.11, R) >droEre2.scaffold_4820 208886 97 - 10470090 ---------CAUAUUCUCGCACUCC-GUGGCCAAAUGCAUUUUGCAUUAAAUUGCAUAACUUUUGUGCGGCAAUUAGAGUU---CUCUGG-CUGGCCAAGACGGUUGCAAA ---------.........(((..((-((((((((((((.....))))).((((((...((....))...))))))(((...---.)))..-.)))))...)))).)))... ( -28.80, z-score = -1.60, R) >droYak2.chr3R 18653919 100 + 28832112 ---------CAUAUUCUUGCACUCC-GUGGCCAAAUGCAUUUUGCAUUAAAUUGCAUAACUUUUGUGCGGCAAUUAGAGCUGCACUCUGG-CUGGCCAAGAAGGCUGGAAG ---------....((((.((.((.(-.((((((.(((((.((((...)))).)))))..((...(((((((.......)))))))...))-.)))))).).)))).)))). ( -34.20, z-score = -2.08, R) >droSec1.super_0 18191083 98 + 21120651 ---------CAUAUUCUCGCACUCCCGUGGCCAAAUGCAUUUUGCAUUAAAUUGCAUAACUUUUGUGCGGCAAUUAGAGUU---CUCUGG-CUGGCCAAGACGGUUGGAAA ---------......(((.(((....)))(((.(((((.....))))).....(((((.....)))))))).....)))..---.((..(-(((.......))))..)).. ( -27.20, z-score = -0.90, R) >droSim1.chr3R 17641224 98 + 27517382 ---------CAUAUUCUCGCACUCCCGUGGCCAAAUGCAUUUUGCAUUAAAUUGCAUAACUUUUGUGCGGCAAUUAGAGUU---CUCUGG-CUGGCCAAGACGGUUGGAAA ---------......(((.(((....)))(((.(((((.....))))).....(((((.....)))))))).....)))..---.((..(-(((.......))))..)).. ( -27.20, z-score = -0.90, R) >consensus _________CAUAUUCUCGCACUCC_GUGGCCAAAUGCAUUUUGCAUUAAAUUGCAUAACUUUUGUGCGGCAAUUAGAGUU___CUCUGG_CUGGCCAAGACGGUUGGAAA ............................((((..((((.....))))..((((((...((....))...))))))..................)))).............. (-13.97 = -14.68 + 0.70)

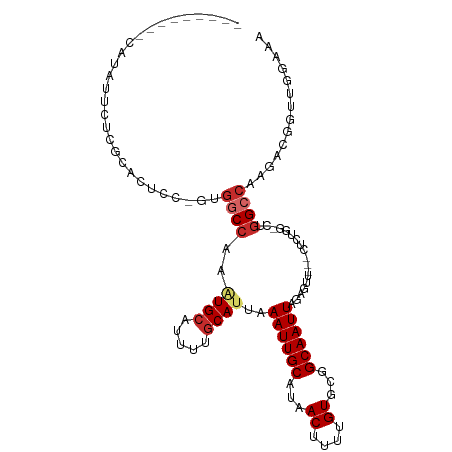
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:49 2011