
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,810,530 – 17,810,620 |
| Length | 90 |
| Max. P | 0.938581 |

| Location | 17,810,530 – 17,810,620 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 89.84 |
| Shannon entropy | 0.17324 |
| G+C content | 0.34989 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -15.88 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
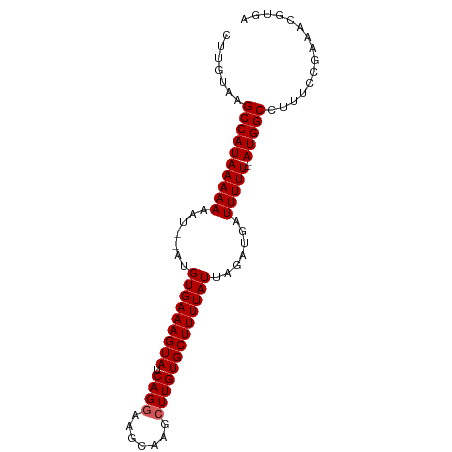
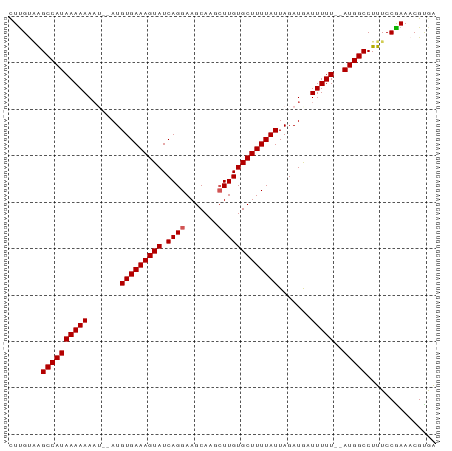
>dm3.chr3R 17810530 90 + 27905053 CUUGCAAGCCAUAAAAAAAGUGAUGUGAAAGUAUCAGGAAGCAAGCUUGUGCUUUUAUUAGAUGAUUUUU--AUGGCCUUUCCAAAACGUGA .......((((((((((..((...(((((((((.((((.......)))))))))))))...))..)))))--)))))............... ( -22.50, z-score = -2.07, R) >droSim1.chr3R 17629037 88 + 27517382 CUUGUGAGCCAUAAAAAAAU--AUGUGAAAGUAUCAGGAAGCAAGCUUGUGCUUUUAUUAGAUGAUUUUU--AUGGCCUUUCCGAAACGUGA .(((.((((((((((((.((--..(((((((((.((((.......)))))))))))))...))..)))))--)))))...)))))....... ( -22.50, z-score = -2.76, R) >droSec1.super_0 18181109 88 + 21120651 CUUGUGAGCCAUAAAAAAAU--AUGUGAAAGUAUCAGGAAGCAAGCUUGUGCUUUUAUUAGAUAAUUUUU--AUGGCCUUUCCGAAACGCGA .(((((.((((((((((...--..(((((((((.((((.......))))))))))))).......)))))--)))))(.....)...))))) ( -21.50, z-score = -2.17, R) >droYak2.chr3R 18643846 89 + 28832112 CUUGUAGGCCAUAAAAAAU---AUGUGAAAGUAUCAGGAAGCAAGCUUGUGCUUUUAUUAGAUGUUUUUUUUAUGGCCUUUCCAAAACGUAG .(((.(((((((((((((.---(((((((((((.((((.......))))))))))))).....)).)))))))))))))...)))....... ( -26.40, z-score = -3.86, R) >droEre2.scaffold_4820 199016 88 - 10470090 UUCGCUGGCCAUAAAAAAAU--AUGUGAAAGUAUCAGGAAGCAAGAUUGUGCUUUUAUUAGAUGUUUUUU--AUGGCCUUUCCGAAACGUAG ((((..((((((((((((..--..(((((((((.(((.........))))))))))))......))))))--))))))....))))...... ( -26.30, z-score = -3.96, R) >consensus CUUGUAAGCCAUAAAAAAAU__AUGUGAAAGUAUCAGGAAGCAAGCUUGUGCUUUUAUUAGAUGAUUUUU__AUGGCCUUUCCGAAACGUGA .......((((((((((.......(((((((((.((((.......))))))))))))).......)))))..)))))............... (-15.88 = -16.08 + 0.20)

| Location | 17,810,530 – 17,810,620 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 89.84 |
| Shannon entropy | 0.17324 |
| G+C content | 0.34989 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -12.34 |
| Energy contribution | -12.30 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
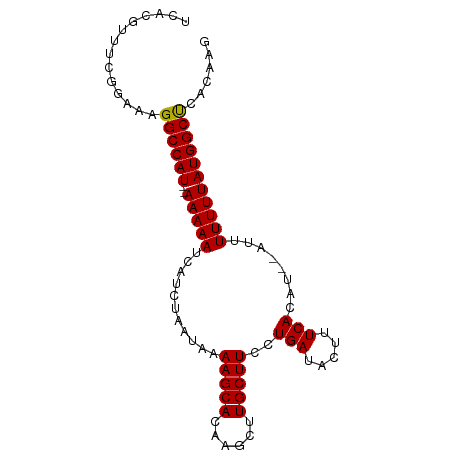
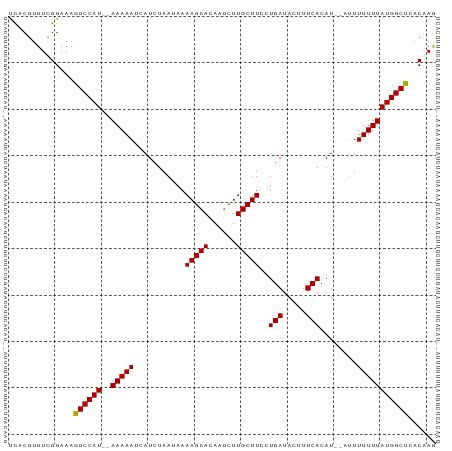
>dm3.chr3R 17810530 90 - 27905053 UCACGUUUUGGAAAGGCCAU--AAAAAUCAUCUAAUAAAAGCACAAGCUUGCUUCCUGAUACUUUCACAUCACUUUUUUUAUGGCUUGCAAG .......(((...(((((((--(((((...........(((((......)))))..((((........))))...)))))))))))).))). ( -18.00, z-score = -2.09, R) >droSim1.chr3R 17629037 88 - 27517382 UCACGUUUCGGAAAGGCCAU--AAAAAUCAUCUAAUAAAAGCACAAGCUUGCUUCCUGAUACUUUCACAU--AUUUUUUUAUGGCUCACAAG ..............((((((--(((((...........(((((......)))))..(((.....)))...--...)))))))))))...... ( -15.80, z-score = -2.45, R) >droSec1.super_0 18181109 88 - 21120651 UCGCGUUUCGGAAAGGCCAU--AAAAAUUAUCUAAUAAAAGCACAAGCUUGCUUCCUGAUACUUUCACAU--AUUUUUUUAUGGCUCACAAG ..............((((((--(((((...........(((((......)))))..(((.....)))...--...)))))))))))...... ( -15.80, z-score = -1.72, R) >droYak2.chr3R 18643846 89 - 28832112 CUACGUUUUGGAAAGGCCAUAAAAAAAACAUCUAAUAAAAGCACAAGCUUGCUUCCUGAUACUUUCACAU---AUUUUUUAUGGCCUACAAG .......(((...(((((((((((((............(((((......)))))..(((.....)))...---.))))))))))))).))). ( -21.60, z-score = -4.14, R) >droEre2.scaffold_4820 199016 88 + 10470090 CUACGUUUCGGAAAGGCCAU--AAAAAACAUCUAAUAAAAGCACAAUCUUGCUUCCUGAUACUUUCACAU--AUUUUUUUAUGGCCAGCGAA ......((((....((((((--((((((..........(((((......)))))..(((.....)))...--..))))))))))))..)))) ( -20.30, z-score = -3.60, R) >consensus UCACGUUUCGGAAAGGCCAU__AAAAAUCAUCUAAUAAAAGCACAAGCUUGCUUCCUGAUACUUUCACAU__AUUUUUUUAUGGCUCACAAG ..............((((((..(((((...........(((((......)))))..(((.....)))......)))))..))))))...... (-12.34 = -12.30 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:47 2011