
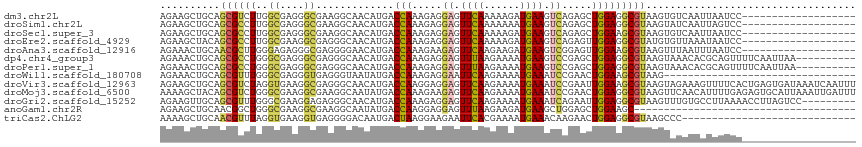
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,219,851 – 8,219,948 |
| Length | 97 |
| Max. P | 0.899540 |

| Location | 8,219,851 – 8,219,948 |
|---|---|
| Length | 97 |
| Sequences | 13 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.66 |
| Shannon entropy | 0.62146 |
| G+C content | 0.46938 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -11.87 |
| Energy contribution | -11.60 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
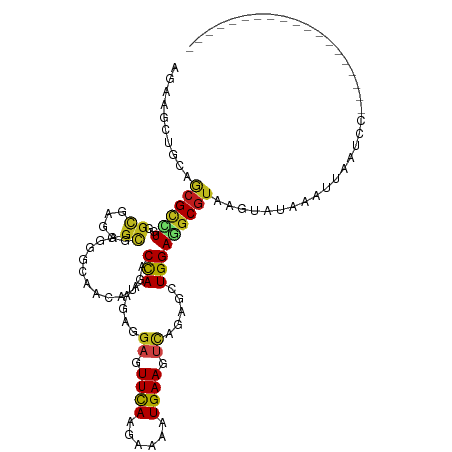
>dm3.chr2L 8219851 97 - 23011544 AGAAGCUGCAGCGUCUUGGCGAGGGCGAAGGCAACAUGACCAAAGAGGAGUUCAAAAAGAUGAAGUCAGAGCUGGAGGCGUAAGUGUCAAUUAAUCC------------------- ........(((((((((.((....)).)))))...............((.((((......)))).))...))))((.((....)).)).........------------------- ( -26.20, z-score = -1.17, R) >droSim1.chr2L 8003628 97 - 22036055 AGAAGCUGCAGCGCCUUGGCGAGGGCGAAGGCAACAUGACCAAAGAGGAGUUCAAAAAAAUGAAGUCAGAGCUGGAGGCGUAAGUAUCAAUUAGUCC------------------- .((..((.(((((((((.((....)).)))))...............((.((((......)))).))...)))).))((....)).)).........------------------- ( -28.40, z-score = -1.90, R) >droSec1.super_3 3717031 97 - 7220098 AGAAGCUGCAGCGCCUUGGCGAGGGCGAAGGCAACAUGACCAAAGAGGAGUUCAAAAAGAUGAAGUCAGAGCUGGAAGCGUAAGUGUCAAUUAAUCC------------------- ....(((.(((((((((.((....)).)))))...............((.((((......)))).))...))))..)))..................------------------- ( -27.60, z-score = -1.55, R) >droEre2.scaffold_4929 17127949 97 - 26641161 AGAAGCUACAGCGCCUUGGCGAAGGCGAGGGCAACAUGACCAAAGAGGAGUUCAAAAAGAUGAAGUCAGAGUUGGAGGCGUAUGUGUUAAAUAAUCC------------------- ....(((((.(((((((.((....)).))))).......((((....((.((((......)))).))....))))..))))).))............------------------- ( -28.40, z-score = -2.81, R) >droAna3.scaffold_12916 6060357 97 - 16180835 AGAAACUGCAACGCUUGGGAGAGGGCGAGGGGAACAUGACCAAAGAAGAGUUCAAGAAGAUGAAGUCGGAGUUGGAAGCGUAAGUUUAAUUUAAUCC------------------- ..(((((...((((((.............(....)....((((....((.((((......)))).))....)))))))))).)))))..........------------------- ( -20.90, z-score = -1.03, R) >dp4.chr4_group3 7707072 106 + 11692001 AGAAACUGCAGCGCCUGGGCGAGGGCGAGGGCAACAUGACCAAAGAGGAGUUUAAGAAAAUGAAGUCCGAGCUGGAGGCGUAAGUAAACACGCAGUUUUCAAUUAA---------- .((((((((.((((((.(((..((.....(....)....)).....(((.((((......)))).)))..))).).)))))..((....)))))))))).......---------- ( -32.50, z-score = -2.22, R) >droPer1.super_1 4826842 106 + 10282868 AGAAACUGCAGCGCCUGGGCGAGGGCGAGGGCAACAUGACCAAAGAGGAGUUUAAGAAAAUGAAGUCCGAGCUGGAGGCGUAAGUAAACACGCAGUUUUCAAUUAA---------- .((((((((.((((((.(((..((.....(....)....)).....(((.((((......)))).)))..))).).)))))..((....)))))))))).......---------- ( -32.50, z-score = -2.22, R) >droWil1.scaffold_180708 2403336 84 - 12563649 AGAAACUGCAGCGUUUGGGCGAGGGUGAGGGUAAUAUGACCAAAGAGGAAUUCAAGAAAAUGAAAUCCGAACUGGAAGCGUAAG-------------------------------- ......(((.((.((..(.....(((.(........).))).....(((.((((......)))).)))...)..)).)))))..-------------------------------- ( -16.10, z-score = -0.67, R) >droVir3.scaffold_12963 10579345 116 + 20206255 AGAAGCUGCAGCGUCUAGGUGAAGGCGAGGGCAACAUGACCAAGGAGGAGUUCAAGAAAAUGAAAUCCGAAUUGGAAGCGUAAGUAGAAAGUUUUCACUGAGUGAUAAAUCAAUUU .((.((....))(((..(((((((((...(....)....((((...(((.((((......)))).)))...))))..((....)).....)))))))))....)))...))..... ( -28.60, z-score = -1.83, R) >droMoj3.scaffold_6500 1633143 116 - 32352404 AAAAGCUACAGCGUCUGGGCGAAGGCGAAGGCAAUAUGACCAAAGAAGAGUUCAAGAAAAUGAAAUCCGAACUGGAGGCGUAAGUUCAACAUUUUGAGAGUGCAUUAAAUUGAUUU ....((....))((((..((....))..))))...................((((...((((...((((((.((..(((....)))...)).)))).))...))))...))))... ( -23.30, z-score = -0.08, R) >droGri2.scaffold_15252 12180546 107 - 17193109 AGAAGUUGCAGCGUUUGGGCGAAGGAGAGGGCAACAUGACCAAAGAGGAGUUCAAGAAAAUGAAAUCAGAAUUGGAGGCGUAAGUUUGUGCCUUAAAACCUUAGUCC--------- ................((((.((((..((((((......((((....((.((((......)))).))....))))((((....)))).))))))....)))).))))--------- ( -27.10, z-score = -1.41, R) >anoGam1.chr2R 58641792 79 - 62725911 AGAAGCUGCAACGGCUGGGCGAAGGCGAAGGCAAUAUGACCAAGGAGGAGUUUAAGAAGAUGAAGCUGGAGCUGGAAGC------------------------------------- ....(((....(((((.(((....((....)).......((.....))................))).).))))..)))------------------------------------- ( -17.00, z-score = -0.28, R) >triCas2.ChLG2 6337656 87 - 12900155 AAAAGCUGCAACGUUUAGGUGAAGGUGAGGGGACAAUGACUAAGGAAGAAUUCACGAAAAUGAAACAAGAACUGGAGGCGUAAGCCC----------------------------- ...........(((((..(((((.....(....).....((.....))..)))))..)))))..............(((....))).----------------------------- ( -14.60, z-score = -0.06, R) >consensus AGAAGCUGCAGCGCCUGGGCGAGGGCGAGGGCAACAUGACCAAAGAGGAGUUCAAGAAAAUGAAGUCAGAGCUGGAGGCGUAAGUAUAAAUUAAUCC___________________ ..........((((((..((....)).............(((.....((.((((......)))).)).....)))))))))................................... (-11.87 = -11.60 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:32 2011