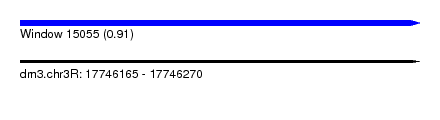
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,746,165 – 17,746,270 |
| Length | 105 |
| Max. P | 0.912549 |

| Location | 17,746,165 – 17,746,270 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.20 |
| Shannon entropy | 0.59358 |
| G+C content | 0.37787 |
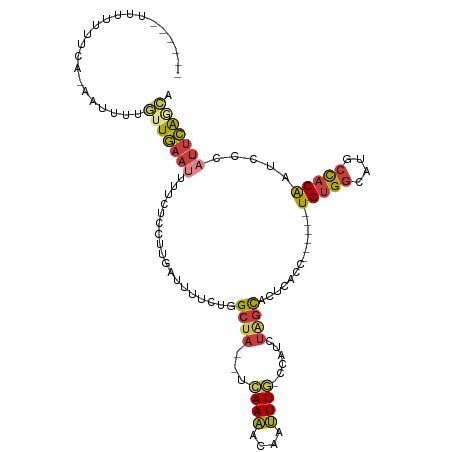
| Mean single sequence MFE | -18.24 |
| Consensus MFE | -10.28 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.16 |
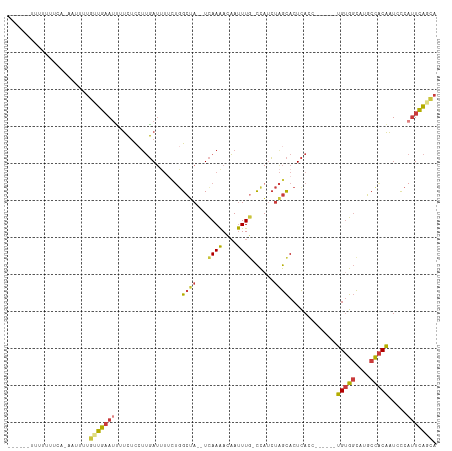
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
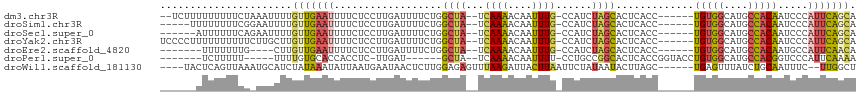
>dm3.chr3R 17746165 105 + 27905053 --UCUUUUUUUUUCUAAAUUUUGUUGAAUUUUCUCCUUGAUUUUCUGGCUA--UCAAAACAAUUUG-CCAUCUAGCACUCACC------UGUGGCAUGCCACAAUCCCAUUCAGCA --...................((((((((................((((..--............)-))).............------(((((....))))).....)))))))) ( -17.74, z-score = -1.75, R) >droSim1.chr3R 17556927 102 + 27517382 -----UUUUUUUUCGGAAUUUUGUUGAAUUUUCUCCUUGAUUUUCUGGCUA--UCAAAACAAUUUG-CCAUCUAGCACUCACC------UGUGGCAUGCCACAAUCCCAUUCAGCA -----................((((((((................((((..--............)-))).............------(((((....))))).....)))))))) ( -17.74, z-score = -0.65, R) >droSec1.super_0 18117217 101 + 21120651 ------AUUUUUUCAGAAUUUUGUUGAAUUUUCUCCUUGAUUUUCUGGCUA--UCAAAACAAUUUG-CCAUCUAGCACUCACC------UGUGGCAUGCCACAAUCCCAUUCAGCA ------...............((((((((................((((..--............)-))).............------(((((....))))).....)))))))) ( -17.74, z-score = -0.97, R) >droYak2.chr3R 18575271 107 + 28832112 UCCCCUUUUUUUUUUCUUGCUUGUUGAAUUUUCUCCUUGAUUUUCUGGCUA--UCAAAACAAUUUG-CCAUCUAGCACUCACC------UGUGGCAUGCCACAAUCCCAUUCAGCA .....................((((((((................((((..--............)-))).............------(((((....))))).....)))))))) ( -17.74, z-score = -1.35, R) >droEre2.scaffold_4820 131601 96 - 10470090 -------UUUUUUUG----CUUGUUGAAUUUUCUCCUUGAUUUUCUGGCUA--UCAAAACAAUUUG-CCAUCUAGCACUCACC------UGUGGCAUGCCACAAUGCCAUUCAACA -------........----..((((((((..((.....))......(((..--...........((-(......)))......------(((((....)))))..))))))))))) ( -19.00, z-score = -1.52, R) >droPer1.super_0 429302 94 + 11822988 -------UCUUUUU-----UUUUGUGCACCACCUC-UUGAU------GCUA--UCAAAACAAUUUU-CCUGCCGGCACUCACCGGUACCUGUGGCAUGCCACGGUCCCAUUCAAAA -------.......-----(((((.(((.((....-.)).)------))..--.))))).......-..((((((......)))))).((((((....))))))............ ( -21.90, z-score = -1.97, R) >droWil1.scaffold_181130 11820569 104 + 16660200 ----UACUCAGUUAAAUGCAUCUAUAAAUAUUAAUGAAUAACUCUUGGAGAGUUUAAGAUUACUUAAUUCUAUAAUACUUAGC------UGAGUUUAUCUGCAAUUUC--UUGGCU ----.(((((((((((((....)))...(((((..(((((((((.....)))))((((....))))))))..))))).)))))------)))))..............--...... ( -15.80, z-score = 0.10, R) >consensus ______UUUUUUUCA_AAUUUUGUUGAAUUUUCUCCUUGAUUUUCUGGCUA__UCAAAACAAUUUG_CCAUCUAGCACUCACC______UGUGGCAUGCCACAAUCCCAUUCAGCA ......................(((((((..................((((...((((....))))......)))).............(((((....))))).....))))))). (-10.28 = -10.57 + 0.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:40 2011