| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,742,616 – 17,742,714 |
| Length | 98 |
| Max. P | 0.773202 |

| Location | 17,742,616 – 17,742,714 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Shannon entropy | 0.45194 |
| G+C content | 0.36322 |
| Mean single sequence MFE | -18.45 |
| Consensus MFE | -8.21 |
| Energy contribution | -9.13 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
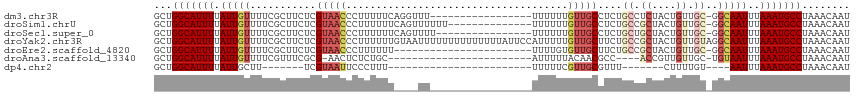
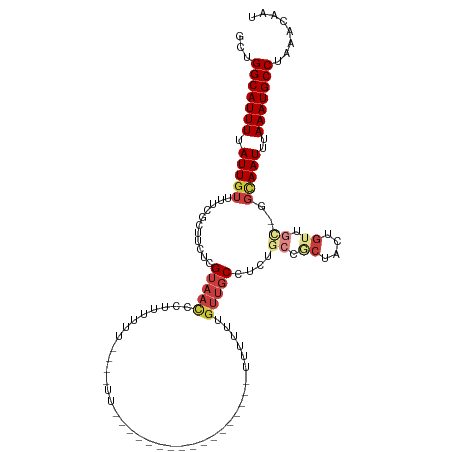
>dm3.chr3R 17742616 98 - 27905053 GCUGGCAUUUUAUUGUUUUCGCUUCUCGUAACCCUUUUUCAGGUUU-----------------UUUUUUGUUGCCUCUGCCUCUACUGUUGC-GGCAAUUUAAAUGCCUAAACAAU ...(((((((.(((((..........((((((........((((..-----------------(.....)..))))...........)))))-))))))..)))))))........ ( -17.31, z-score = -1.02, R) >droSim1.chrU 6442958 101 + 15797150 GCUGGCAUUUUAUUGUUUUCGCUUCUCGUAACCCUUUUUUCAGUUUUUU--------------UUUUUUGUUGCCUCUGCCGCUACUGUUGC-GGCAAUUUAAAUGCCUAAACAAU ...(((((((.................(((((.................--------------......)))))...((((((.......))-))))....)))))))........ ( -20.30, z-score = -2.15, R) >droSec1.super_0 18113694 99 - 21120651 GCUGGCAUUUUAUUGUUUUCGCUUCUCGUAACCCUUUUUUCAGUUUU----------------UUUUUUGUUGCCUCUGCUGCUACUGUUGC-GGCAAUUUAAAUGCCUAAACAAU ...(((((((.................(((((...............----------------......)))))...((((((.......))-))))....)))))))........ ( -18.20, z-score = -1.35, R) >droYak2.chr3R 18571599 116 - 28832112 GCUGGCAUUUUAUUGUUUUCGCUUCUCGUAACCCUUUUUUGUAAUUUUUUUUUUUUUUAUUCCAUUUUUGUUGCUUCUGCCGCUACUGUUGUAGGCAAUUUAAAUGCCUAAACAAU ...(((((((.(((((...........(((((........((((............)))).........)))))..((((.((....)).)))))))))..)))))))........ ( -19.53, z-score = -1.98, R) >droEre2.scaffold_4820 128195 92 + 10470090 GCUGGCAUUUUAUUGUUUUCGCUUCUCGUAACCCUUUUUU-----------------------UUUUGUGUUGCUUCUGCCGCUACUGUUGC-GGCAAUUUAAAUGCCUAAACAAU ...(((((((.................(((((.(......-----------------------....).)))))...((((((.......))-))))....)))))))........ ( -22.30, z-score = -3.21, R) >droAna3.scaffold_13340 8749621 86 - 23697760 GCUGGCAUUUUAUUGUUUUCGUUUCGCG-AACUCUCUGC------------------------AUUUUUACAACGCC----ACCGUUGUUGC-UGUAAUUUAAAUGCCUAAACAAU ...(((((((.((((..((((.....))-))......((------------------------(.....((((((..----..)))))))))-..))))..)))))))........ ( -18.00, z-score = -1.93, R) >dp4.chr2 6600049 74 - 30794189 GCUGGCAUUUUAUUGCUU-------UCGUAAUUCCCUUU------------------------UUUUUCGUUGCGUUU-------CUUUUGU----AAUUUAAAUGCCUAAACAAU ...(((((((.(((((..-------.((((((.......------------------------......))))))...-------.....))----)))..)))))))........ ( -13.52, z-score = -3.03, R) >consensus GCUGGCAUUUUAUUGUUUUCGCUUCUCGUAACCCUUUUUU____UU_________________UUUUUUGUUGCCUCUGCCGCUACUGUUGC_GGCAAUUUAAAUGCCUAAACAAU ...(((((((....(((............))).....................................((((((...((.((....)).)).))))))..)))))))........ ( -8.21 = -9.13 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:39 2011