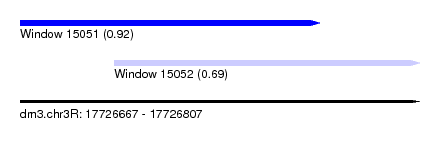
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,726,667 – 17,726,807 |
| Length | 140 |
| Max. P | 0.921796 |

| Location | 17,726,667 – 17,726,772 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.69 |
| Shannon entropy | 0.41970 |
| G+C content | 0.41414 |
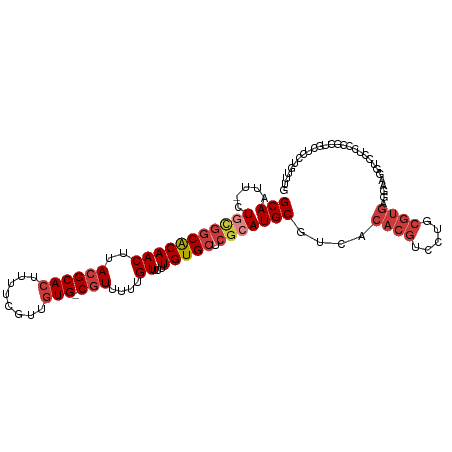
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.71 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
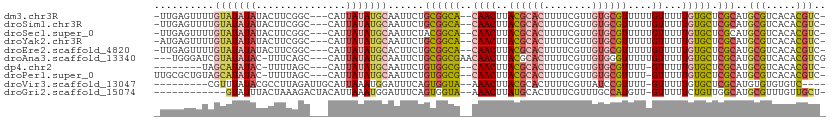
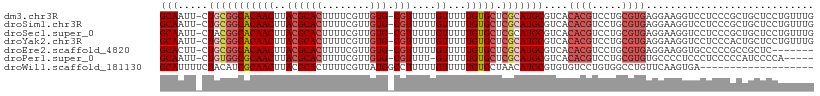
>dm3.chr3R 17726667 105 + 27905053 -UUGAGUUUUGUAUAUAUACUUCGGC---CAUUAUAUGCAAUUCUGCGGCA--CAACUUACGCACUUUUCGUUGUGCGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUC- -...((..(((((((((..(....).---...)))))))))..))((((((--((((..((((((........))))))....))...))))).)))..(((.....))).- ( -32.40, z-score = -3.33, R) >droSim1.chr3R 17537821 105 + 27517382 -UUGAGUUUUGUAUAUAUACUUCGGC---CAUUAUAUGCAAUUCUGCGGCA--CAACUUACGCACUUUUCGUUGUGCGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUC- -...((..(((((((((..(....).---...)))))))))..))((((((--((((..((((((........))))))....))...))))).)))..(((.....))).- ( -32.40, z-score = -3.33, R) >droSec1.super_0 18097852 105 + 21120651 -UUGAGUUUUGUAUAUAUACUUCGGC---CAUUAUAUGCAAUUCUACGGCA--CAACUUACGCACUUUUCGUUGUGCGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUC- -...((..(((((((((..(....).---...)))))))))..)).(((((--((((..((((((........))))))....))...))))).))...(((.....))).- ( -28.20, z-score = -2.42, R) >droYak2.chr3R 18555079 105 + 28832112 -AUGAGUUUUGUAUAUAUACUUCGGC---CAUUAUAUGCAAUUCUGCGGCA--CAACUUACGCACUUUUCGUUGUGCGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUC- -...((..(((((((((..(....).---...)))))))))..))((((((--((((..((((((........))))))....))...))))).)))..(((.....))).- ( -32.40, z-score = -3.29, R) >droEre2.scaffold_4820 112389 105 - 10470090 -UUGAGUUUUGUAUAUAUACUUCGGC---CAUUAUAUGCACUUCUGCGGCA--CAACUUACGCACUUUUCGUUGUGCGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUC- -........((((((((..(....).---...))))))))....(((((((--((((..((((((........))))))....))...))))).)))).(((.....))).- ( -29.60, z-score = -2.44, R) >droAna3.scaffold_13340 8725476 105 + 23697760 ---UGGGAUCGUAUAUAC-UUUCAGC---CAUUAUAUGCAAUUCUGCGGCGAACAACUUACGCACUUUUCGUUGUGGGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUCG ---.(((((.(((((((.-.......---...))))))).))))).((((((((((...((.(((........))).))..)))))..((((..((....)).))))))))) ( -25.40, z-score = -0.42, R) >dp4.chr2 6583609 96 + 30794189 --------UAGCAUAUAC-UUUUAGC---CAUUAUAUGCAAUUCUGUGGCG--CAACUUACGCACUUUUCGUUGUGCGUUUU-GUUUUUGUGCUCGCAUGCGUCACACGUC- --------..(((((((.-.......---...))))))).....(((((((--((((..((((((........))))))...-)))...(((....)))))))))))....- ( -30.00, z-score = -3.07, R) >droPer1.super_0 409379 104 + 11822988 UUGCGCUGUAGCAUAUAC-UUUUAGC---CAUUAUAUGCAAUUCUGUGGCG--CAACUUACGCACUUUUCGUUGUGCGUUUU-GUUUUUGUGCUCGCAUGCGUCACACGUC- ..(((.....(((((((.-.......---...))))))).....(((((((--((((..((((((........))))))...-)))...(((....)))))))))))))).- ( -31.00, z-score = -1.75, R) >droVir3.scaffold_13047 10455746 96 - 19223366 ---------CGUUUAUACGCCUUAGAUUGCAUUAAAUGGAUUUCAGUGGUA--AAACUUACGCACUUUUCGUUAUCCGUUUU-GUUUUUGUGCUCGCAUGUGUGUGUC---- ---------....(((((((....((..(((..((((((((..((((((((--.....))).))))....)..)))))))).-.....)))..))....)))))))..---- ( -20.50, z-score = -0.95, R) >droGri2.scaffold_15074 863270 96 + 7742996 ------------GUAUUUACUAAAGACUACAUUAAAUGGAUUUCAGUGGUA--AAACUUAUGCACUUUUCGUUUGCCAUGUU-GUUUUUCUGUUGGCAUGCGUUUGUUGCU- ------------...(((((((..((...(((...)))....))..)))))--))......(((.....(((.(((((....-..........))))).))).....))).- ( -17.84, z-score = -0.07, R) >consensus _UUGAGUUUUGUAUAUAUACUUCAGC___CAUUAUAUGCAAUUCUGCGGCA__CAACUUACGCACUUUUCGUUGUGCGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUC_ ..........(((((((...............)))))))......((((((...(((..((((((........))))))....)))....))).)))..(((.....))).. (-15.28 = -15.71 + 0.43)
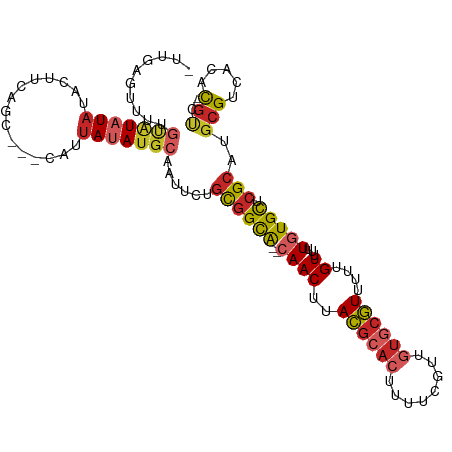
| Location | 17,726,700 – 17,726,807 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Shannon entropy | 0.36198 |
| G+C content | 0.52569 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -17.38 |
| Energy contribution | -18.64 |
| Covariance contribution | 1.27 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17726700 107 + 27905053 GCAAUU-CUGCGGCACAACUUACGCACUUUUCGUUGUG-CGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUCCUGCGUGAGGAAGGUCCUCCCGCUGCUCCUGUUUG (((...-.(((((((((((..((((((........)))-)))....))...))))).)))))))...(((.(..(.(((.((((.....)))).))).)..).)))... ( -36.30, z-score = -2.52, R) >droSim1.chr3R 17537854 107 + 27517382 GCAAUU-CUGCGGCACAACUUACGCACUUUUCGUUGUG-CGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUCCUGCGUGAGGAAGGUCCUCCCGCUGCUCCUGUUUG (((...-.(((((((((((..((((((........)))-)))....))...))))).)))))))...(((.(..(.(((.((((.....)))).))).)..).)))... ( -36.30, z-score = -2.52, R) >droSec1.super_0 18097885 107 + 21120651 GCAAUU-CUACGGCACAACUUACGCACUUUUCGUUGUG-CGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUCCUGCGUGAGGAAGGUCCUCCCGCUGCUCCUGUUUG (((...-....(((((((...((((((........)))-)))..))))...((((..((....)).)))).)))..(((.((((.....)))).))))))......... ( -31.10, z-score = -1.34, R) >droYak2.chr3R 18555112 107 + 28832112 GCAAUU-CUGCGGCACAACUUACGCACUUUUCGUUGUG-CGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUCCUGCGUGAGGAAGGUCCUCCCACUGCUCCUGUUUG (((...-.(((((((((((..((((((........)))-)))....))...))))).)))))))....((((.....))))((((.(((......)))..))))..... ( -34.40, z-score = -2.27, R) >droEre2.scaffold_4820 112422 100 - 10470090 GCACUU-CUGCGGCACAACUUACGCACUUUUCGUUGUG-CGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUCCUGCGUGAGGAAGGUGCCCCCGCCGCUC------- ((((((-((((((((((((..((((((........)))-)))....))...))))).)))........((((.....)))).)))).))))...........------- ( -34.20, z-score = -1.49, R) >droPer1.super_0 409412 101 + 11822988 GCAAUU-CUGUGGCGCAACUUACGCACUUUUCGUUGUG-CGUUUU-GUUUUUGUGCUCGCAUGCGUCACACGUCCUGCGUGUGCCCCUCCCUCCCCCAUCCCCA----- (((...-.(((((((((((..((((((........)))-)))...-))...))))).)))))))(.((((((.....)))))))....................----- ( -30.90, z-score = -4.36, R) >droWil1.scaffold_181130 11798365 90 + 16660200 GCAUUUUCUACAUCGCAACUUACGCACUUUUCGUUAUGGCCUUUUUGUUUUUGUGCUAACAUGCGUGUGUCCUGUGGCCUGUUCAAGUGA------------------- .(((((...(((..((.((..((((((....(((..(((((...........).))))..))).))))))...)).)).)))..))))).------------------- ( -15.00, z-score = 1.02, R) >consensus GCAAUU_CUGCGGCACAACUUACGCACUUUUCGUUGUG_CGUUUUUGUUUUUGUGCUCGCAUGCGUCACACGUCCUGCGUGAGGAAGGUCCUCCCGCUGCUCCUGUUUG (((.....(((((((((((..((((((........))).)))....))...))))).)))))))....((((.....))))............................ (-17.38 = -18.64 + 1.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:37 2011