
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,677,215 – 17,677,316 |
| Length | 101 |
| Max. P | 0.500000 |

| Location | 17,677,215 – 17,677,316 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.84 |
| Shannon entropy | 0.42919 |
| G+C content | 0.36288 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -8.73 |
| Energy contribution | -10.10 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 17677215 101 - 27905053 CCCCGCCAUACACAGAUACUUUUGUGUAUGGAUUGUACGUUUGUUUUAAACCAAGACUUGACGGUCCAAAAAUGUUGUUGCAU-AACAAAUGAUUAAGUGAA .....(((((((((((....)))))))))))......((((((((.........((((....)))).....((((....))))-)))))))).......... ( -25.30, z-score = -2.37, R) >droSim1.chr3R 17486076 101 - 27517382 CCCAGCCAUACACAGAGAGUAUUGUGUAUGGAUUGUACGUUUGUUUUAAACCAAGACUUGACGGUCCCAAAAUGUUGUUGCAU-AACAAAUGAUUAAGCGAA ..(((((((((((((......)))))))))).)))..((((((((.........((((....)))).....((((....))))-)))))))).......... ( -23.90, z-score = -1.85, R) >droSec1.super_0 18048704 101 - 21120651 CCCAGCCAUACACAGAGAGUAUUGUGUAUGGAUUGUACGUUUGUUUUAAACCAAGACUUAACGGUCCCAAAAUGUUGUUGCAU-AACAAAUGAUUAAGUGAA ..(((((((((((((......)))))))))).)))..((((((((.........((((....)))).....((((....))))-)))))))).......... ( -23.90, z-score = -2.02, R) >droYak2.chr3R 18502565 98 - 28832112 CCCCGCCAUACACAGAUGCAUUUGUGUAUGGAUCGUACGUUUGUUUUAAACCAAGACUU----GUCCCAAAAUGUUGUUGCAUAAAAAAAUGAUUAAGUGAA ...(((((((((((((....))))))))))((((((......(((((.....))))).(----((..(((....)))..))).......))))))..))).. ( -20.00, z-score = -1.45, R) >droEre2.scaffold_4820 59713 101 + 10470090 CCCCGCCAUACACAGAUACCCUUGUGUAUGGAUUGUAAGUUUGUUUUAAACCAAGACUUGACGGUCCCAAAAUGUUGUUGCAU-AAAAAAUGAUUAAGUGAA ..(((((((((((((......))))))))))....(((((((...........))))))).)))...((....((..((....-....))..))....)).. ( -21.90, z-score = -2.00, R) >dp4.chr2 6532546 90 - 30794189 ---UUACGUAUGCAUAAAGCUUU---UAUG---UAUAUAAUUG-UUUAAACAAAGACUUGACGAUCCC-AAAUGUUGUCCAAC-AAAAAGCGCCAAGUCGAA ---....((((((((((.....)---))))---)))))..(((-(....)))).((((((.((.....-...((((....)))-).....)).))))))... ( -18.92, z-score = -2.89, R) >droPer1.super_0 358103 90 - 11822988 ---UUACGUAUGCAUAAAGCUUU---UAUG---UAUAUAAUUG-UUUAAACAAAGACUUGACGGUCCC-AAAUGUUGUCCAAC-AAAAAGCGCCAAGUCGAA ---....((((((((((.....)---))))---)))))..(((-(....)))).((((((.((.(...-...((((....)))-)...).)).))))))... ( -18.50, z-score = -2.02, R) >consensus CCCCGCCAUACACAGAUAGCUUUGUGUAUGGAUUGUACGUUUGUUUUAAACCAAGACUUGACGGUCCCAAAAUGUUGUUGCAU_AAAAAAUGAUUAAGUGAA .....((((((((((......)))))))))).......................(((......))).................................... ( -8.73 = -10.10 + 1.37)

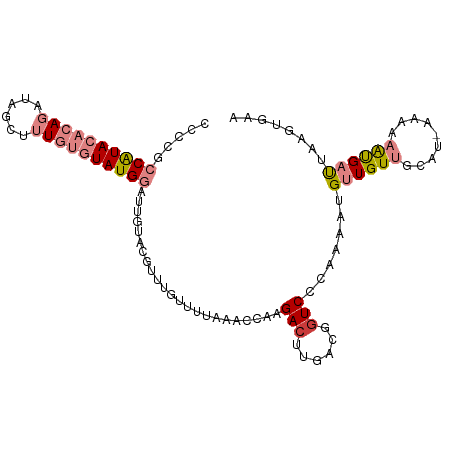
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:30 2011