
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,663,850 – 17,663,984 |
| Length | 134 |
| Max. P | 0.776648 |

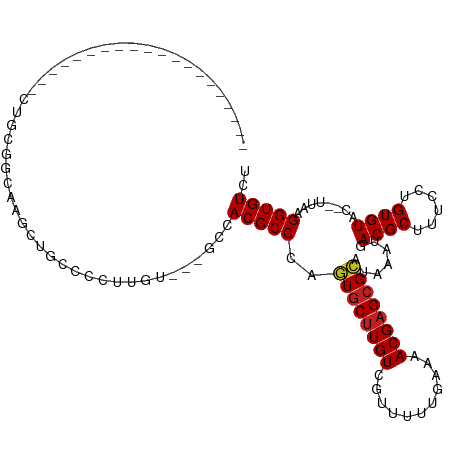
| Location | 17,663,850 – 17,663,956 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.34 |
| Shannon entropy | 0.52230 |
| G+C content | 0.53489 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
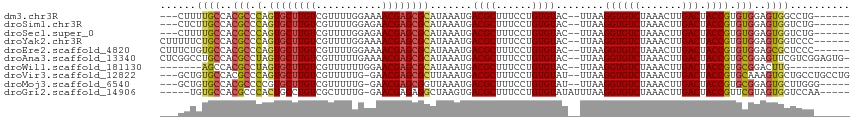
>dm3.chr3R 17663850 106 + 27905053 ------CCCCCCCUCCUGCCCAGCGGCAAGCUGCCCCUUUU---GCCACGCCCAGUGCUUGUCGUUUUGGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCU ------..................((((((........)))---)))(((((..((((((((..((...))..)))))))).......((((......))))..--....))))).. ( -26.10, z-score = -0.21, R) >droSim1.chr3R 17470923 106 + 27517382 ------CCACCCCUCUUUGCCCGCGGCAAGCUGCCCCUCUU---GCCACGCCCAGUGCUUGUCGUUUUGGAGAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCU ------.((((......((..((.((((((........)))---))).))..))(((((((((........).)))))))).......((((......))))..--....))))... ( -30.30, z-score = -1.08, R) >droSec1.super_0 18036610 106 + 21120651 ------CCCACCCUCUUUGCCUGCGGCAAGCUGCCCCUUUU---GCCACGCCCAGUGCUUGUCGUUUUGGAGAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCU ------..................((((((........)))---)))(((((..(((((((((........).)))))))).......((((......))))..--....))))).. ( -27.30, z-score = -0.12, R) >droYak2.chr3R 18486652 111 + 28832112 ---CCCCUCCCUCACCUUCCCUGCUGCA-GCUGCCCCUUUUUCUGCCACGCCCAGUGCUUGUCGUUUUGGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCU ---.........((((((....((.(((-(............))))...))...((((((((..((...))..)))))))).......((((......))))..--..))))))... ( -22.50, z-score = -0.32, R) >droEre2.scaffold_4820 48839 114 - 10470090 CGGCUAACCCUCCCCCUGGCCUGCGGCG-GCUGCCCCUUUCUGUGCCACGCCCAGUGCUUGUCGUUUUGGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCU .(((((..........)))))((.((((-...((.(......).))..))))))((((((((..((...))..))))))))......((((((((...(....)--..)))))))). ( -31.00, z-score = 0.53, R) >droAna3.scaffold_13340 8660413 104 + 23697760 -----------CCACCCCCCACUGUCCGGCCUGCCCCUCGGCCUGCCACGCCUAGUGCUUGUCGUUUUUGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCU -----------................(((..(((....)))..)))((((((.((((((((..((...))..)))))))).......((((......))))..--...)))))).. ( -30.10, z-score = -2.43, R) >droPer1.super_0 344020 92 + 11822988 --------------------CCCCUCCGAACUACCCGUC---CUGCCACGCCUAGUGCUUGUCGUUUUUGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCU --------------------...................---.....((((((.((((((((..((...))..)))))))).......((((......))))..--...)))))).. ( -20.80, z-score = -1.69, R) >dp4.chr2 6518426 92 + 30794189 --------------------CCCCUCCGAACUACCCGUC---CUGCCACGCCUAGUGCUUGUCGUUUUUGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCU --------------------...................---.....((((((.((((((((..((...))..)))))))).......((((......))))..--...)))))).. ( -20.80, z-score = -1.69, R) >droWil1.scaffold_181130 11719556 95 + 16660200 --------------------CCUUUUUUUGUUUUUCUUGGCUAAGCCACGCCUAGUGCUUGUCGUUUUUUGGAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCU --------------------..................(((...)))((((((.(((((((((........).)))))))).......((((......))))..--...)))))).. ( -24.10, z-score = -1.52, R) >droVir3.scaffold_12822 3973407 91 + 4096053 --------------------AUGCAGCAGUCUGCGGGCUGU---GCCACGCCCAGUGCUUGUCGUUUUUG-GAACGAGCGCUUAAAUGACGCUUUCCUGUGUAU--UUAAGGUGUCU --------------------..(((.(((((....))))))---)).(((((..(((((((((.......-).))))))))(((((((.(((......))))))--))))))))).. ( -30.50, z-score = -1.12, R) >droMoj3.scaffold_6540 17249603 91 + 34148556 --------------------CUGGGCCUGACGGCAGGCUGU---GCCACGCCCCGUGCUUGUCGUUUUUG-GAACGAGCGGUUAAAUGACGCUUUCCUGUGUAU--UUAAGGUGUCU --------------------...((((((....))))))..---...(((((((((.....((((((...-))))))))))(((((((.(((......))))))--))))))))).. ( -27.90, z-score = 0.03, R) >droGri2.scaffold_14906 1528212 90 + 14172833 --------------------CUGGGGCAGACUG---AGUGU---GCCACGCCCACUGCCUGUCGCUUUUG-GAACGAGAGGCUAAGUGACGCUUUCCUGUGUAUAUUUAAGGUGUCU --------------------....((((.((..---.)).)---)))(((((((((((((.(((......-...))).))))..))))((((......))))........))))).. ( -29.60, z-score = -0.91, R) >consensus ____________________CUGCGGCAAGCUGCCCCUUGU___GCCACGCCCAGUGCUUGUCGUUUUUGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC__UUAAGGUGUCU ...............................................(((((..((((((((...........)))))))).......((((......))))........))))).. (-17.57 = -17.68 + 0.11)


| Location | 17,663,880 – 17,663,984 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.03 |
| Shannon entropy | 0.33648 |
| G+C content | 0.49737 |
| Mean single sequence MFE | -30.91 |
| Consensus MFE | -21.69 |
| Energy contribution | -21.60 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17663880 104 + 27905053 ---CUUUUGCCACGCCCAGUGCUUGUCGUUUUGGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCUAAACUUGACUACCGUGUGGAGUGGCCUG------ ---.....(((((.((((((((((((..((...))..)))))))).......((((......))))..--....((((((.......))).))).)).)).)))))...------ ( -32.20, z-score = -2.17, R) >droSim1.chr3R 17470953 104 + 27517382 ---CUCUUGCCACGCCCAGUGCUUGUCGUUUUGGAGAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCUAAACUUGACUACCGUGUGGAGUGGUCUG------ ---.....(((((.(((((((((((((........).)))))))).......((((......))))..--....((((((.......))).))).)).)).)))))...------ ( -31.50, z-score = -1.55, R) >droSec1.super_0 18036640 104 + 21120651 ---CUUUUGCCACGCCCAGUGCUUGUCGUUUUGGAGAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCUAAACUUGACUACCGUGUGGAGUGGUCUG------ ---.....(((((.(((((((((((((........).)))))))).......((((......))))..--....((((((.......))).))).)).)).)))))...------ ( -31.50, z-score = -1.66, R) >droYak2.chr3R 18486684 107 + 28832112 CUUUUUCUGCCACGCCCAGUGCUUGUCGUUUUGGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCUAAACUUGACUACCGUGUGGAGUGGUCCC------ ........(((((.((((((((((((..((...))..)))))))).......((((......))))..--....((((((.......))).))).)).)).)))))...------ ( -30.30, z-score = -1.55, R) >droEre2.scaffold_4820 48874 107 - 10470090 CUUUCUGUGCCACGCCCAGUGCUUGUCGUUUUGGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCUAAACUUGACUACCGUGUGGAGCGCUCCC------ ......((.((((((...((((((((..((...))..)))))))).......((((......))))..--....((((((.......))).))))))))).))......------ ( -30.60, z-score = -1.55, R) >droAna3.scaffold_13340 8660438 112 + 23697760 CUCGGCCUGCCACGCCUAGUGCUUGUCGUUUUUGAAAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCUAAACUUGACUACCGUGCGGAGUUCGUCGGAGUG- ((((((.......)))..((((((((..((...))..)))))))).....(((((..((((...((((--....((((((.......))).)))))))))))..))))))))..- ( -33.10, z-score = -0.91, R) >droWil1.scaffold_181130 11719579 96 + 16660200 -------AGCCACGCCUAGUGCUUGUCGUUUUUUGGAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC--UUAAGGUGUCUAAACUUGACUACCGUGCGGACUUG---------- -------..((.(((...(((((((((........).)))))))).......((((......))))..--....((((((.......))).)))))).)).....---------- ( -25.50, z-score = -0.84, R) >droVir3.scaffold_12822 3973423 109 + 4096053 ---GCUGUGCCACGCCCAGUGCUUGUCGUUUUUG-GAACGAGCGCUUAAAUGACGCUUUCCUGUGUAU--UUAAGGUGUCUAAACUUGACUACCGUGCAAAGUGCUGCCUGCCUG ---.......((.((.(((..(((.((((((...-))))))(((((((((((.(((......))))))--))))((((((.......))).))))))).)))..)))...)).)) ( -31.70, z-score = -1.66, R) >droMoj3.scaffold_6540 17249619 104 + 34148556 ---GCUGUGCCACGCCCCGUGCUUGUCGUUUUUG-GAACGAGCGGUUAAAUGACGCUUUCCUGUGUAU--UUAAGGUGUCUAAACUUGACUACCGUGCGGAGUGCUUGGG----- ---......(((.((((((..(...((((((...-))))))....(((((((.(((......))))))--))))((((((.......))).))))..))).).)).))).----- ( -31.00, z-score = -0.67, R) >droGri2.scaffold_14906 1528227 104 + 14172833 -----UGUGCCACGCCCACUGCCUGUCGCUUUUG-GAACGAGAGGCUAAGUGACGCUUUCCUGUGUAUAUUUAAGGUGUCUAAACUUGACUACCGUUCGUAGUGGUCCAA----- -----((..((((...((((((((.(((......-...))).))))..))))((((......))))........((((((.......))).))).......))))..)).----- ( -31.70, z-score = -1.76, R) >consensus ___CUUGUGCCACGCCCAGUGCUUGUCGUUUUGGAGAACGAGCGCAUAAAUGACGCUUUCCUGUGUAC__UUAAGGUGUCUAAACUUGACUACCGUGCGGAGUGGUCCG______ ......((((..(((.(.((((((((...........)))))))).......((((......))))........((((((.......))).)))).)))..)))).......... (-21.69 = -21.60 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:28 2011