
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,196,324 – 8,196,424 |
| Length | 100 |
| Max. P | 0.872240 |

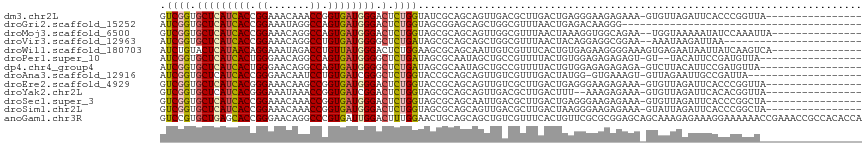
| Location | 8,196,324 – 8,196,424 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.71 |
| Shannon entropy | 0.67216 |
| G+C content | 0.50961 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.28 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8196324 100 + 23011544 GUCGGUGCUCAUCACCGGAAACAAACCGGUGAUGGGACUCUGGUAUCGCAGCAGUUGACGCUUGACUGAGGGAAGAGAAA-GUGUUAGAUUCACCCGGUUA---------------- ...(((((((((((((((.......)))))))))))((((((......))).)))((((((((..((......))...))-))))))....))))......---------------- ( -34.40, z-score = -1.58, R) >droGri2.scaffold_15252 5095499 77 - 17193109 AUCGGUGCUCAUCACCGGAAAUAGGCCAGUGAUGGGACUCUGGUAGCGGAGCAGCUGGCGUUUAACUGAGACAAGGG---------------------------------------- .((((((.....))))))......((((((.....(.(((((....)))))).))))))((((.....)))).....---------------------------------------- ( -24.10, z-score = -0.71, R) >droMoj3.scaffold_6500 30801869 100 - 32352404 GUCGGUGCUCAUCACCGGAAACAGGCCAGUGAUGGGACUCUGGUAGCGCAGCAGUUGGCGUUUAACUAAAGGUGGCAGAA--UGGUAAAAAUAUCCAAAUUA--------------- ((((...((((((((.((.......)).)))))))).((.(((((((((........)))))..)))).)).))))....--(((.........))).....--------------- ( -25.60, z-score = -0.03, R) >droVir3.scaffold_12963 9707980 92 + 20206255 AUCGGUGCUCAUCACCGGAAACAGGCCUGUGAUGGGGCUCUGAUAGCGCAGCAGCUGGCGUUUAACUACAGGAGGCGGAA--AAAUAAGAUUAA----------------------- .((((.(((((((((.((.......)).))))).)))).))))((((......))))((.(((.......))).))....--............----------------------- ( -24.40, z-score = 0.23, R) >droWil1.scaffold_180703 2867410 102 + 3946847 AUCUGUACUCAUAACAGGAAAUAGACCUGUUAUGGGACUCUGGAAGCGCAGCAAUUGUCGUUUCACUGUGAGAAGGGGAAAGUGAGAAUAAUUAUCAAGUCA--------------- .(((...(((((((((((.......))))))))))).(((.((((((((((...))).)))))).)...))).))).((...(((.........)))..)).--------------- ( -26.60, z-score = -1.29, R) >droPer1.super_10 634865 97 + 3432795 AUCGGUGCUCAUCACUGGGAACAGGCCAGUGAUGGGGCUCUGAUAGCGCAAUAGCUGCCGUUUUACUGUGGAGAGAGAGU-GU--UACAUUCCGAUGUUA----------------- (((((..(((((((((((.......)))))))))))(((((..((((......))))(((........)))....)))))-..--......)))))....----------------- ( -33.60, z-score = -1.51, R) >dp4.chr4_group4 1631475 99 + 6586962 AUCGGUGCUCAUCACUGGGAACAGGCCAGUGAUGGGGCUCUGAUAGCGCAAUAGCUGCCGUUUUACUGUGGAGAGAGAGA-GUCUUACAUUCCGAUGUUA----------------- (((((..(((((((((((.......)))))))))))(((((..((((......))))(((........)))......)))-))........)))))....----------------- ( -33.00, z-score = -1.06, R) >droAna3.scaffold_12916 9541066 96 + 16180835 AUCGGUGCUCAUCACCGGGAACAAUCCUGUGAUCGGGCUCUGGUACCGCAGCAGUUGUCGUUUGACUAUGG-GUGAAAGU-GUUAGAAUUGCCGAUUA------------------- (((((((((((((((.((((....))))))))).))))(((..(((..((.(....(((....)))....)-.))...))-)..)))...))))))..------------------- ( -32.30, z-score = -1.54, R) >droEre2.scaffold_4929 17102377 100 + 26641161 GUCGGUGCUCAUCACGGGAAACAAGCCGGUGAUGGGACUCUGGUACCGCAGCAGUUGUCGCUUGACUGAGGGAAGAGAAA-GUGUUAGAUUCACCCGGUUA---------------- ...((((((((((((.((.......)).)))))))).((((....((....((((((.....)))))).))..))))...-..........))))......---------------- ( -31.30, z-score = -0.13, R) >droYak2.chr2L 10828385 98 + 22324452 GUCGGUGCUCAUCACGGGAAAUAAACCGGUGAUCGGACUCUGGUAGCGCAGCAGUUGACGCUUGACUUU--AAAGAGAAA-GUGUUAGAUUCACACGGUUA---------------- .((.(((.....))).)).....(((((((((((.(((((((......))).))))(((((((..(((.--...))).))-))))).)).)))).))))).---------------- ( -27.50, z-score = -0.76, R) >droSec1.super_3 3693283 100 + 7220098 GUCGGUGCUCAUCACCGGAAACAAACCGGUGAUGGGACUCUGGUAGCGCAGCAAUUGACGCUUGACUGAGGGAAGAGAAA-GUGUUAGAUUCACCCGGCUA---------------- (((((..(((((((((((.......))))))))))).((((((((((((((...))).))))..))).))))........-(((.......))))))))..---------------- ( -35.70, z-score = -2.06, R) >droSim1.chr2L 7979395 100 + 22036055 GUCGGUGCUCAUCACCGGAAACAAACCGGUGAUGGGACUCUGGUAGCGCAGCAGUUGACGCUUGACUAAGGGAAGAGAAA-GUAUUAGAUUCACCCGGCUA---------------- ((((((((((((((((((.......))))))))))).((((((((((((((...))).))))..))).))))........-..........)).)))))..---------------- ( -35.30, z-score = -2.26, R) >anoGam1.chr3R 30915493 117 + 53272125 GUCCGUGCUGAGCACCGGGAACAGGCCCGUGAUUGGACUUUGGAACUGCAGCAGCUGUCGUUUCACUGUUCGCGCGGAGCAGCAAAGAGAAAGGAAAAAACCGAAACCGCCACACCA .((..(((((..(..((.((((((..(((....)))....((((((.((((...)))).)))))))))))).))..)..)))))..))....((......))............... ( -35.40, z-score = 0.32, R) >consensus GUCGGUGCUCAUCACCGGAAACAGGCCGGUGAUGGGACUCUGGUAGCGCAGCAGUUGACGUUUGACUGAGGGAAGAGAAA_GUGUUAAAUUCACACGGUU_________________ .((((.(((((((((.((.......)).)))))))).).)))).......................................................................... (-15.33 = -15.28 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:30 2011