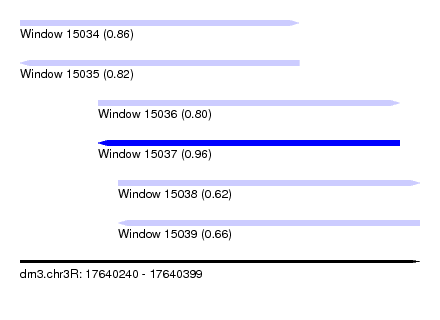
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,640,240 – 17,640,399 |
| Length | 159 |
| Max. P | 0.962862 |

| Location | 17,640,240 – 17,640,351 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.15 |
| Shannon entropy | 0.10755 |
| G+C content | 0.29804 |
| Mean single sequence MFE | -22.01 |
| Consensus MFE | -15.83 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17640240 111 + 27905053 UCCUAUACUUUUUCAUUUAAAAUAUUGUAUAUAUAUUACAAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUGAAAGCACUUUGAUCCCAACGAUAUAUGGUUACAAA ..(((((.................((((((((.........))))))))........((((((((((...(....)....)))))))))).........)))))....... ( -17.80, z-score = -1.42, R) >droSec1.super_0 18012524 109 + 21120651 GCACAUACUUCUUCAUUUAAAAUA--GUGUAUAUAUUACAAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUCGAACCACUUUGAUCCCGACGAUAUAUGGUUACAAA ...((((..((.((.......(((--((((((((.......)))))).)))))....((((((((((.............))))))))))..)).))..))))........ ( -24.42, z-score = -3.55, R) >droSim1.chr3R 17445812 109 + 27517382 GCAUAUACUUCUUCAUUUAAAAUA--UUGUAUAUAUUACAAAUAUGCAACUUUAAAAGGUCAAAGUGAAGCCUUUCGAAGCACUUUGAUCCCGACGAUAUAUGGUUACAAA .((((((..((.((.((((((...--((((((((.......)))))))).)))))).((((((((((...(.....)...))))))))))..)).))))))))........ ( -23.80, z-score = -3.26, R) >consensus GCAUAUACUUCUUCAUUUAAAAUA__GUGUAUAUAUUACAAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUCGAAGCACUUUGAUCCCGACGAUAUAUGGUUACAAA ...((((..((.((.............(((((((.......))))))).........((((((((((.............))))))))))..)).))..))))........ (-15.83 = -16.05 + 0.23)

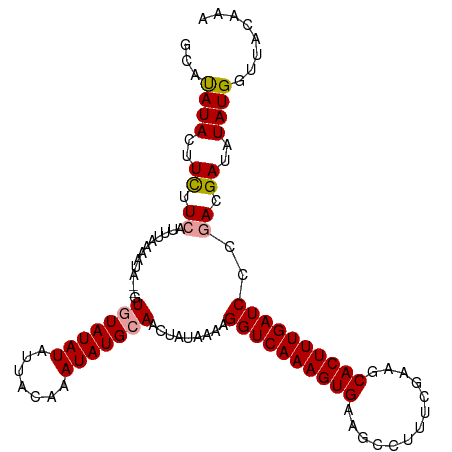
| Location | 17,640,240 – 17,640,351 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.15 |
| Shannon entropy | 0.10755 |
| G+C content | 0.29804 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -20.72 |
| Energy contribution | -20.17 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.815761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17640240 111 - 27905053 UUUGUAACCAUAUAUCGUUGGGAUCAAAGUGCUUUCAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUUGUAAUAUAUAUACAAUAUUUUAAAUGAAAAAGUAUAGGA .......((((((((....(((.(((((((((((....))))...))))))))))......((((((....))))))))))))...(((((((.......))))))).)). ( -20.30, z-score = -0.95, R) >droSec1.super_0 18012524 109 - 21120651 UUUGUAACCAUAUAUCGUCGGGAUCAAAGUGGUUCGAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUUGUAAUAUAUACAC--UAUUUUAAAUGAAGAAGUAUGUGC ........(((((.((.(((((.(((((((((.((....))..))))))))))))..((((((((((....)))))).))))...--..........)).)).)))))... ( -26.20, z-score = -2.35, R) >droSim1.chr3R 17445812 109 - 27517382 UUUGUAACCAUAUAUCGUCGGGAUCAAAGUGCUUCGAAAGGCUUCACUUUGACCUUUUAAAGUUGCAUAUUUGUAAUAUAUACAA--UAUUUUAAAUGAAGAAGUAUAUGC ........(((((((..(((((.((((((((..((....))...)))))))))))......((((((....))))))........--.............)).))))))). ( -24.00, z-score = -2.21, R) >consensus UUUGUAACCAUAUAUCGUCGGGAUCAAAGUGCUUCGAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUUGUAAUAUAUACAC__UAUUUUAAAUGAAGAAGUAUAUGC ........(((((((..(((((.((((((((..((....))...)))))))))))......((((((....)))))).......................)).))))))). (-20.72 = -20.17 + -0.55)

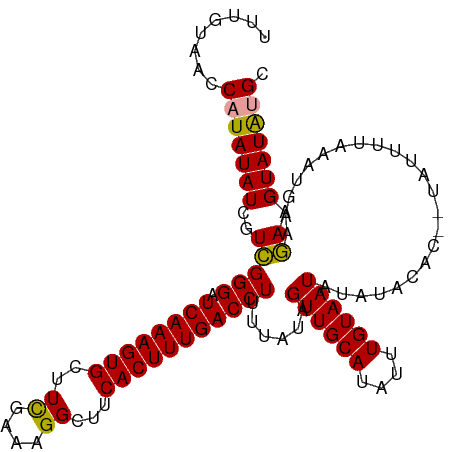
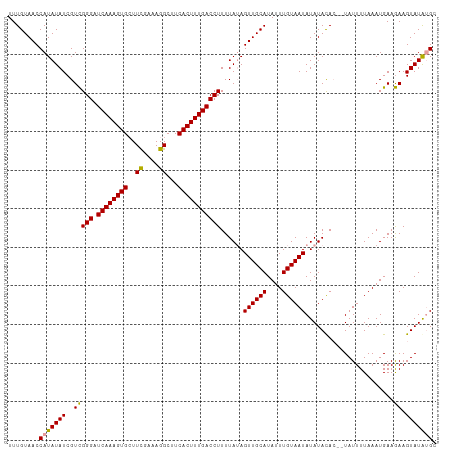
| Location | 17,640,271 – 17,640,391 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Shannon entropy | 0.03826 |
| G+C content | 0.36111 |
| Mean single sequence MFE | -22.74 |
| Consensus MFE | -21.89 |
| Energy contribution | -21.89 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17640271 120 + 27905053 UAUAUUACAAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUGAAAGCACUUUGAUCCCAACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUUUGCCAUUACUGCCACCA ((((((.....((((....))))...((((((((((...(....)....)))))))))).....))))))((((....((((.(((((..((....))..))))).))))...))))... ( -22.70, z-score = -1.42, R) >droSec1.super_0 18012553 120 + 21120651 UAUAUUACAAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUCGAACCACUUUGAUCCCGACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUUUGCCAUUACUGCCACCA ((((((.....((((....))))...((((((((((.............)))))))))).....))))))((((....((((.(((((..((....))..))))).))))...))))... ( -22.72, z-score = -1.68, R) >droSim1.chr3R 17445841 120 + 27517382 UAUAUUACAAAUAUGCAACUUUAAAAGGUCAAAGUGAAGCCUUUCGAAGCACUUUGAUCCCGACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUUUGCCAUUACUGCCACCA ..........................((((((((((...(.....)...))))))))))...........((((....((((.(((((..((....))..))))).))))...))))... ( -22.80, z-score = -1.33, R) >consensus UAUAUUACAAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUCGAAGCACUUUGAUCCCGACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUUUGCCAUUACUGCCACCA ..........................((((((((((.............))))))))))...........((((....((((.(((((..((....))..))))).))))...))))... (-21.89 = -21.89 + -0.00)

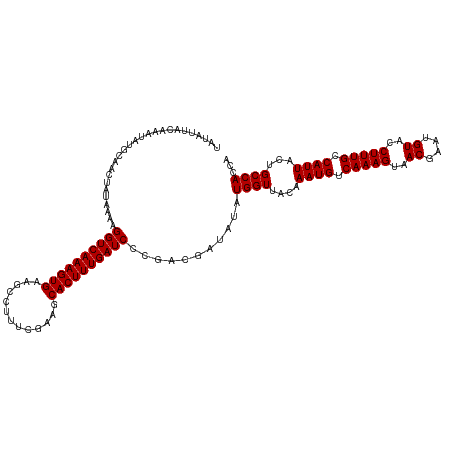
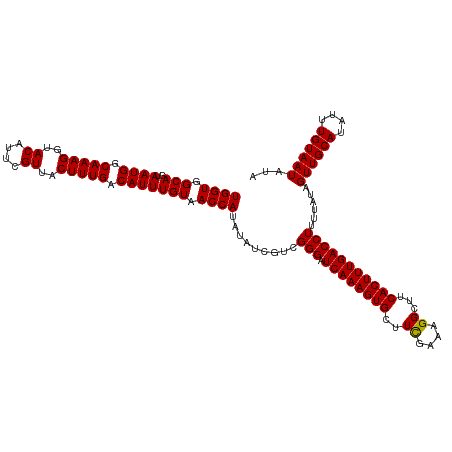
| Location | 17,640,271 – 17,640,391 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Shannon entropy | 0.03826 |
| G+C content | 0.36111 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -30.09 |
| Energy contribution | -29.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17640271 120 - 27905053 UGGUGGCAGUAAUGGCAAAGGUACAUUCGUUACUUUGACAUUUGUAACCAUAUAUCGUUGGGAUCAAAGUGCUUUCAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUUGUAAUAUA ((((.(((..((((.(((((..((....))..))))).))))))).))))((((.....(((.(((((((((((....))))...))))))))))..))))((((((....))))))... ( -29.50, z-score = -1.61, R) >droSec1.super_0 18012553 120 - 21120651 UGGUGGCAGUAAUGGCAAAGGUACAUUCGUUACUUUGACAUUUGUAACCAUAUAUCGUCGGGAUCAAAGUGGUUCGAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUUGUAAUAUA ((((.(((..((((.(((((..((....))..))))).))))))).)))).........(((.(((((((((.((....))..))))))))))))......((((((....))))))... ( -33.60, z-score = -2.80, R) >droSim1.chr3R 17445841 120 - 27517382 UGGUGGCAGUAAUGGCAAAGGUACAUUCGUUACUUUGACAUUUGUAACCAUAUAUCGUCGGGAUCAAAGUGCUUCGAAAGGCUUCACUUUGACCUUUUAAAGUUGCAUAUUUGUAAUAUA ((((.(((..((((.(((((..((....))..))))).))))))).)))).........(((.((((((((..((....))...)))))))))))......((((((....))))))... ( -31.90, z-score = -2.29, R) >consensus UGGUGGCAGUAAUGGCAAAGGUACAUUCGUUACUUUGACAUUUGUAACCAUAUAUCGUCGGGAUCAAAGUGCUUCGAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUUGUAAUAUA ((((.(((..((((.(((((..((....))..))))).))))))).)))).........(((.((((((((..((....))...)))))))))))......((((((....))))))... (-30.09 = -29.87 + -0.22)

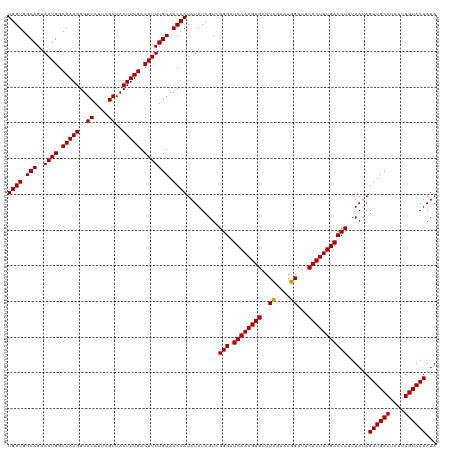
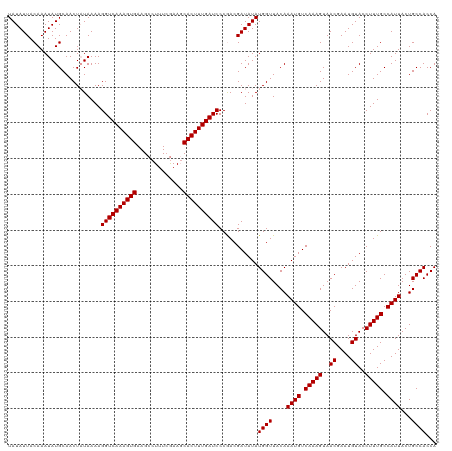
| Location | 17,640,279 – 17,640,399 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Shannon entropy | 0.04591 |
| G+C content | 0.40000 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -22.63 |
| Energy contribution | -22.41 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17640279 120 + 27905053 AAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUGAAAGCACUUUGAUCCCAACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUUUGCCAUUACUGCCACCACCCACUGG ..................((((((((((...(....)....))))))))))(((........((((....((((.(((((..((....))..))))).))))...))))........))) ( -22.99, z-score = -1.08, R) >droSec1.super_0 18012561 120 + 21120651 AAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUCGAACCACUUUGAUCCCGACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUUUGCCAUUACUGCCACCAUCCACUGG ..................((((((((((.............))))))))))(((..(((...((((....((((.(((((..((....))..))))).))))...))))..)))...))) ( -23.52, z-score = -1.32, R) >droSim1.chr3R 17445849 120 + 27517382 AAAUAUGCAACUUUAAAAGGUCAAAGUGAAGCCUUUCGAAGCACUUUGAUCCCGACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUUUGCCAUUACUGCCACCACCCACUGG ..................((((((((((...(.....)...))))))))))...........((((....((((.(((((..((....))..))))).))))...))))(((.....))) ( -23.30, z-score = -0.92, R) >consensus AAAUAUGCAACUAUAAAAGGUCAAAGUGAAGCCUUUCGAAGCACUUUGAUCCCGACGAUAUAUGGUUACAAAUGUCAAAGUAACGAAUGUACCUUUGCCAUUACUGCCACCACCCACUGG ..................((((((((((.............))))))))))(((........((((....((((.(((((..((....))..))))).))))...))))........))) (-22.63 = -22.41 + -0.22)

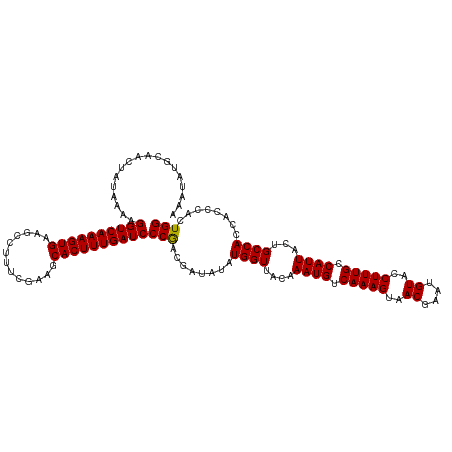
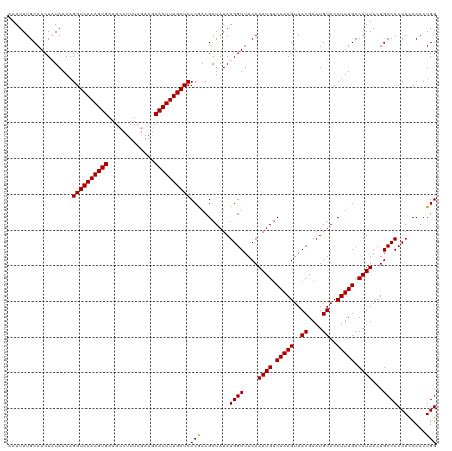
| Location | 17,640,279 – 17,640,399 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Shannon entropy | 0.04591 |
| G+C content | 0.40000 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -29.91 |
| Energy contribution | -29.47 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.660279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17640279 120 - 27905053 CCAGUGGGUGGUGGCAGUAAUGGCAAAGGUACAUUCGUUACUUUGACAUUUGUAACCAUAUAUCGUUGGGAUCAAAGUGCUUUCAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUU ...(..((((((.(((..((((.(((((..((....))..))))).))))))).)))))........(((.(((((((((((....))))...)))))))))).......)..)...... ( -28.40, z-score = -0.43, R) >droSec1.super_0 18012561 120 - 21120651 CCAGUGGAUGGUGGCAGUAAUGGCAAAGGUACAUUCGUUACUUUGACAUUUGUAACCAUAUAUCGUCGGGAUCAAAGUGGUUCGAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUU ...(..((((((.(((..((((.(((((..((....))..))))).))))))).)))))........(((.(((((((((.((....))..)))))))))))).......)..)...... ( -33.00, z-score = -1.93, R) >droSim1.chr3R 17445849 120 - 27517382 CCAGUGGGUGGUGGCAGUAAUGGCAAAGGUACAUUCGUUACUUUGACAUUUGUAACCAUAUAUCGUCGGGAUCAAAGUGCUUCGAAAGGCUUCACUUUGACCUUUUAAAGUUGCAUAUUU ...(..((((((.(((..((((.(((((..((....))..))))).))))))).)))))........(((.((((((((..((....))...))))))))))).......)..)...... ( -31.60, z-score = -1.34, R) >consensus CCAGUGGGUGGUGGCAGUAAUGGCAAAGGUACAUUCGUUACUUUGACAUUUGUAACCAUAUAUCGUCGGGAUCAAAGUGCUUCGAAAGGCUUCACUUUGACCUUUUAUAGUUGCAUAUUU ...(..((((((.(((..((((.(((((..((....))..))))).))))))).)))))........(((.((((((((..((....))...))))))))))).......)..)...... (-29.91 = -29.47 + -0.44)

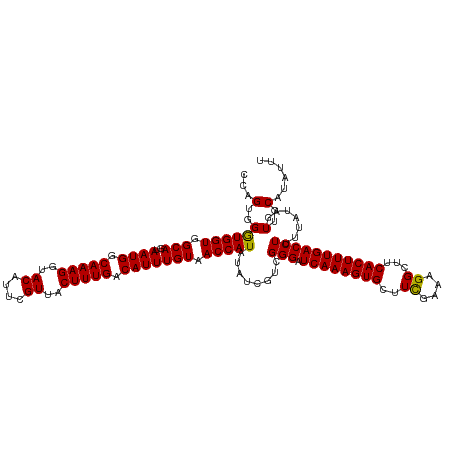
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:26 2011