
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,623,947 – 17,624,029 |
| Length | 82 |
| Max. P | 0.812488 |

| Location | 17,623,947 – 17,624,029 |
|---|---|
| Length | 82 |
| Sequences | 10 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 91.18 |
| Shannon entropy | 0.17242 |
| G+C content | 0.46110 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.69 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
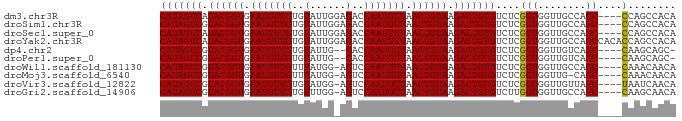
>dm3.chr3R 17623947 82 - 27905053 CAUAGUCAUACGGUGAAUGUUGUGUAUUGGAGACCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUGCCAGC----CCAGCCACA (((((((.((((((.(((((((.((.......))))))))).)))))).))))))).....(((((((.....----.))))))). ( -30.80, z-score = -3.10, R) >droSim1.chr3R 17429599 82 - 27517382 CAUAGUCGUACGGUGAAUGUUGUGUAUUGGAGACCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUGCCAGC----CCAGCCACA (((((((.((((((.(((((((.((.......))))))))).)))))).))))))).....(((((((.....----.))))))). ( -30.80, z-score = -3.08, R) >droSec1.super_0 17996315 82 - 21120651 CAUAGUCAUACGGUGAAUGUUGUGUAUUGGAGACCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUGCCAGC----CCAGCCACA (((((((.((((((.(((((((.((.......))))))))).)))))).))))))).....(((((((.....----.))))))). ( -30.80, z-score = -3.10, R) >droYak2.chr3R 18443935 86 - 28832112 CAUAGUCAUACGGUGAAUGUUGUGUAUUGGAGACCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUGCCAGCCACACCAGCCACA (((((((.((((((.(((((((.((.......))))))))).)))))).))))))).....(((((((..........))))))). ( -29.70, z-score = -1.93, R) >dp4.chr2 6480715 79 - 30794189 CAUAGUCGUACGGUGAAUGUUGUGUAUUG--GACCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUGUCAGC----CAAGCAGC- (((((((.((((((.(((((((.((....--.))))))))).)))))).)))))))......((((.....))----))......- ( -24.70, z-score = -2.07, R) >droPer1.super_0 306015 79 - 11822988 CAUAGUCGUACGGUGAAUGUUGUGUAUUG--GACCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUGUCAGC----CAAGCAGC- (((((((.((((((.(((((((.((....--.))))))))).)))))).)))))))......((((.....))----))......- ( -24.70, z-score = -2.07, R) >droWil1.scaffold_181130 11672711 81 - 16660200 CAUAGUCGUACGGUGAAUGUUGUUUAUGG-AGUCCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUGCCAGC----CAAACAACA (((((((.((((((.(((((((..(....-.)..))))))).)))))).)))))))....(((((...))).)----)........ ( -23.10, z-score = -1.43, R) >droMoj3.scaffold_6540 17209646 80 - 34148556 CAUAGUCGUACGGUGAAUGUUGUUUAUGG-AGUCCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUG-CAGC----CAAACAACA (((((((.((((((.(((((((..(....-.)..))))))).)))))).)))))))......((((..-..))----))....... ( -23.10, z-score = -1.61, R) >droVir3.scaffold_12822 3936741 81 - 4096053 CAUAGUCGUACGGUGAAUGUUGUGUAUGG-AGUCCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUGUUAGC----UAAUCAACA (((((((.((((((.(((((((.(.....-...)))))))).)))))).)))))))....(.((((((.....----)))))).). ( -22.50, z-score = -1.97, R) >droGri2.scaffold_14906 1491731 81 - 14172833 CAUAGUCGUACGGUGAAUGUUGUGUUUGG-AGUCCAAUGUUAACUGUAAGACUGUGUCUUGGUGGUUGCCAGC----CAAGCAACA (((((((.((((((.(((((((..(....-.)..))))))).)))))).))))))).((((((((...))).)----))))..... ( -27.10, z-score = -2.53, R) >consensus CAUAGUCGUACGGUGAAUGUUGUGUAUUG_AGACCAAUGUUAACUGUAAGACUGUGUCUCGGUGGUUGCCAGC____CAAGCAACA (((((((.((((((.(((((((..(......)..))))))).)))))).)))))))....(((....)))................ (-19.86 = -19.69 + -0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:19 2011