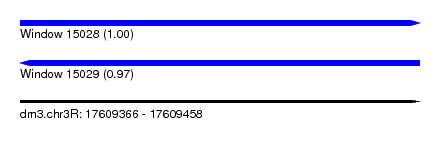
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,609,366 – 17,609,458 |
| Length | 92 |
| Max. P | 0.998898 |

| Location | 17,609,366 – 17,609,458 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 67.86 |
| Shannon entropy | 0.64047 |
| G+C content | 0.36587 |
| Mean single sequence MFE | -16.60 |
| Consensus MFE | -8.02 |
| Energy contribution | -8.32 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.998898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
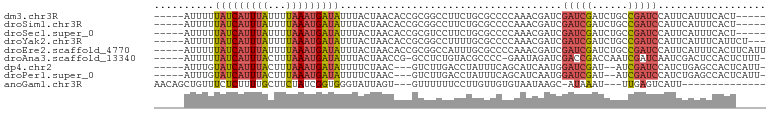
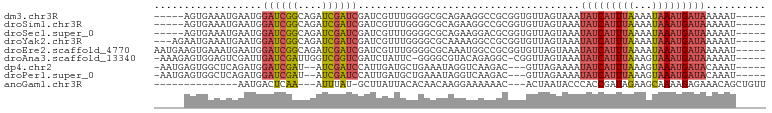
>dm3.chr3R 17609366 92 + 27905053 -----AUUUUUAUCAUUUAUUUUAAAUGAUAUUUACUAACACCGCGGCCUUCUGCGCCCCAAACGAUCGAUCGAUCUGCCGAUCCAUUCAUUUCACU----- -----.....(((((((((...)))))))))...........(((((....)))))........((..(((((......)))))...))........----- ( -16.60, z-score = -2.80, R) >droSim1.chr3R 17415059 92 + 27517382 -----AUUUUUAUCAUUUAUUUUAAAUGAUAUUUACUAACACCGCGGCCUUCUGCGCCCCAAACGAUCGAUCGAUCUGCCGAUCCAUUCAUUUCACU----- -----.....(((((((((...)))))))))...........(((((....)))))........((..(((((......)))))...))........----- ( -16.60, z-score = -2.80, R) >droSec1.super_0 17981583 92 + 21120651 -----AUUUUUAUCAUUUAUUUUAAAUGAUAUUUACUAACACCGCGUCCUUCUGCGCCCCAAACGAUCGAUCGAUCUGCCGAUCCAUUCAUUUCACU----- -----.....(((((((((...)))))))))............((((......)))).......((..(((((......)))))...))........----- ( -15.70, z-score = -3.30, R) >droYak2.chr3R 18425916 94 + 28832112 -----AUUUUUAUCAUUUAUUUUAAAUGAUAUUUACUAACACCGCGGCCUUUUGCGCCCCAAACGAUCGAUCGAUCUGCCGAUCCAUUCAUUUCAUUCU--- -----.....(((((((((...)))))))))............(.((((....).))).)....((..(((((......)))))...))..........--- ( -15.80, z-score = -2.22, R) >droEre2.scaffold_4770 17738739 97 + 17746568 -----AUUUUUAUCAUUUAUUUUAAAUGAUAUUUACUAACACCGCGGCCAUUUGCGCCCCAAACGAUCGAUCGAUCUGCCGAUCCAUUCAUUUCACUUCAUU -----.....(((((((((...)))))))))............(.((((....).))).)....((..(((((......)))))...))............. ( -15.60, z-score = -2.15, R) >droAna3.scaffold_13340 8599385 94 + 23697760 -----AUUUUUAUCAUUUACUUUAAAUGAUAUUUACUAACCG-GCCUCUGUACGCCCC-GAAUAGAUCGACCGACCAAUCGAUCAAUCGACUCCACUCUUU- -----.....(((((((((...)))))))))..........(-((........))).(-((...((((((........))))))..)))............- ( -16.70, z-score = -3.12, R) >dp4.chr2 6466751 91 + 30794189 -----AUUUGUAUCAUUUACUUUAAAUGAUAUUUUCUAAC---GUCUUGACCUAUUUCAGCAUCAAUGGAUCGAU--AUCGAUCCAUCUGAGCCACUCAUU- -----....((((((((((...))))))))))........---................((.(((((((((((..--..)))))))).)))))........- ( -21.70, z-score = -3.99, R) >droPer1.super_0 292609 91 + 11822988 -----AUUUGUAUCAUUUACUUUAAAUGAUAUUUUCUAAC---GUCUUGACCUAUUUCAGCAUCAAUGGAUCGAU--AUCGAUCCAUCUGAGCCACUCAUU- -----....((((((((((...))))))))))........---................((.(((((((((((..--..)))))))).)))))........- ( -21.70, z-score = -3.99, R) >anoGam1.chr3R 40625984 81 - 53272125 AACAGCUGUUUCUCUUUUGCUUCUAUCGGUGGGUAUUAGU---GUUUUUUCCUUGUUGUGUAAUAAGC-AUAAAU---UUGAGUCAUU-------------- ......((...(((....(((..((((....))))..)))---............((((((.....))-))))..---..))).))..-------------- ( -9.00, z-score = 0.97, R) >consensus _____AUUUUUAUCAUUUAUUUUAAAUGAUAUUUACUAACACCGCGUCCUUCUGCGCCCCAAACGAUCGAUCGAUCUGCCGAUCCAUUCAUUUCACU_____ ..........(((((((((...))))))))).....................................(((((......))))).................. ( -8.02 = -8.32 + 0.30)
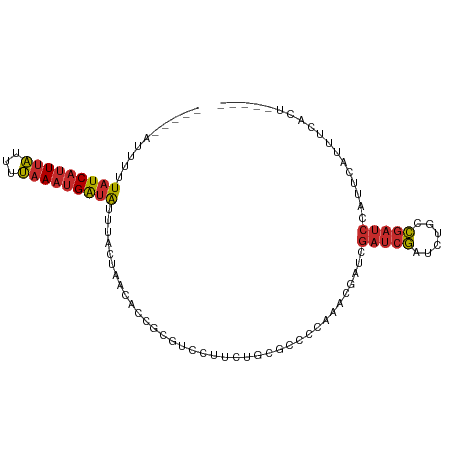
| Location | 17,609,366 – 17,609,458 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 67.86 |
| Shannon entropy | 0.64047 |
| G+C content | 0.36587 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -8.76 |
| Energy contribution | -9.02 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
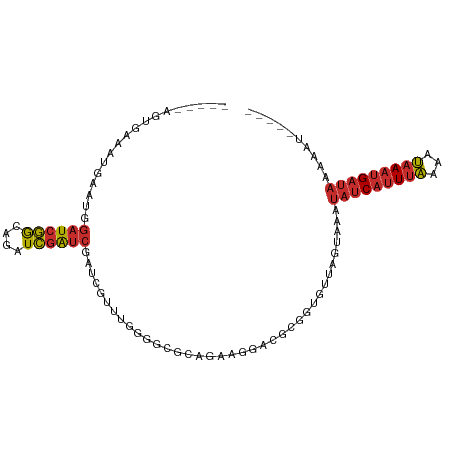
>dm3.chr3R 17609366 92 - 27905053 -----AGUGAAAUGAAUGGAUCGGCAGAUCGAUCGAUCGUUUGGGGCGCAGAAGGCCGCGGUGUUAGUAAAUAUCAUUUAAAAUAAAUGAUAAAAAU----- -----....((((((.(.((((((....)))))).)))))))(.(((.......))).)............(((((((((...))))))))).....----- ( -21.50, z-score = -1.54, R) >droSim1.chr3R 17415059 92 - 27517382 -----AGUGAAAUGAAUGGAUCGGCAGAUCGAUCGAUCGUUUGGGGCGCAGAAGGCCGCGGUGUUAGUAAAUAUCAUUUAAAAUAAAUGAUAAAAAU----- -----....((((((.(.((((((....)))))).)))))))(.(((.......))).)............(((((((((...))))))))).....----- ( -21.50, z-score = -1.54, R) >droSec1.super_0 17981583 92 - 21120651 -----AGUGAAAUGAAUGGAUCGGCAGAUCGAUCGAUCGUUUGGGGCGCAGAAGGACGCGGUGUUAGUAAAUAUCAUUUAAAAUAAAUGAUAAAAAU----- -----..(.((((((.(.((((((....)))))).))))))).)(.(((........))).).........(((((((((...))))))))).....----- ( -19.30, z-score = -1.36, R) >droYak2.chr3R 18425916 94 - 28832112 ---AGAAUGAAAUGAAUGGAUCGGCAGAUCGAUCGAUCGUUUGGGGCGCAAAAGGCCGCGGUGUUAGUAAAUAUCAUUUAAAAUAAAUGAUAAAAAU----- ---......((((((.(.((((((....)))))).)))))))(.(((.......))).)............(((((((((...))))))))).....----- ( -21.50, z-score = -1.70, R) >droEre2.scaffold_4770 17738739 97 - 17746568 AAUGAAGUGAAAUGAAUGGAUCGGCAGAUCGAUCGAUCGUUUGGGGCGCAAAUGGCCGCGGUGUUAGUAAAUAUCAUUUAAAAUAAAUGAUAAAAAU----- .........((((((.(.((((((....)))))).)))))))(.(((.......))).)............(((((((((...))))))))).....----- ( -21.50, z-score = -1.24, R) >droAna3.scaffold_13340 8599385 94 - 23697760 -AAAGAGUGGAGUCGAUUGAUCGAUUGGUCGGUCGAUCUAUUC-GGGGCGUACAGAGGC-CGGUUAGUAAAUAUCAUUUAAAGUAAAUGAUAAAAAU----- -...(((((((.((((((((((....)))))))))))))))))-..(((........))-)..........(((((((((...))))))))).....----- ( -30.30, z-score = -4.66, R) >dp4.chr2 6466751 91 - 30794189 -AAUGAGUGGCUCAGAUGGAUCGAU--AUCGAUCCAUUGAUGCUGAAAUAGGUCAAGAC---GUUAGAAAAUAUCAUUUAAAGUAAAUGAUACAAAU----- -.......(((...(((((((((..--..)))))))))...)))...............---.........(((((((((...))))))))).....----- ( -22.80, z-score = -2.84, R) >droPer1.super_0 292609 91 - 11822988 -AAUGAGUGGCUCAGAUGGAUCGAU--AUCGAUCCAUUGAUGCUGAAAUAGGUCAAGAC---GUUAGAAAAUAUCAUUUAAAGUAAAUGAUACAAAU----- -.......(((...(((((((((..--..)))))))))...)))...............---.........(((((((((...))))))))).....----- ( -22.80, z-score = -2.84, R) >anoGam1.chr3R 40625984 81 + 53272125 --------------AAUGACUCAA---AUUUAU-GCUUAUUACACAACAAGGAAAAAAC---ACUAAUACCCACCGAUAGAAGCAAAAGAGAAACAGCUGUU --------------.....(((..---.(((.(-((((............((.......---........))..(....))))))))))))........... ( -3.56, z-score = 1.14, R) >consensus _____AGUGAAAUGAAUGGAUCGGCAGAUCGAUCGAUCGUUUGGGGCGCAGAAGGACGCGGUGUUAGUAAAUAUCAUUUAAAAUAAAUGAUAAAAAU_____ ..................((((((....)))))).....................................(((((((((...))))))))).......... ( -8.76 = -9.02 + 0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:18 2011