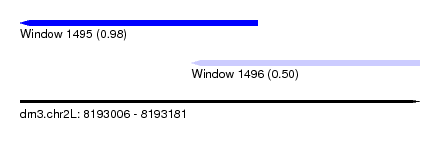
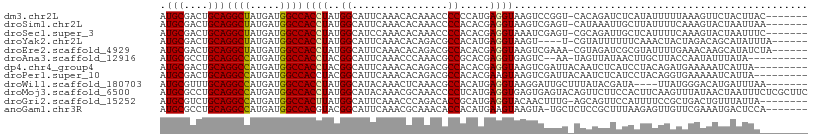
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,193,006 – 8,193,181 |
| Length | 175 |
| Max. P | 0.984060 |

| Location | 8,193,006 – 8,193,110 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 62.96 |
| Shannon entropy | 0.79286 |
| G+C content | 0.40274 |
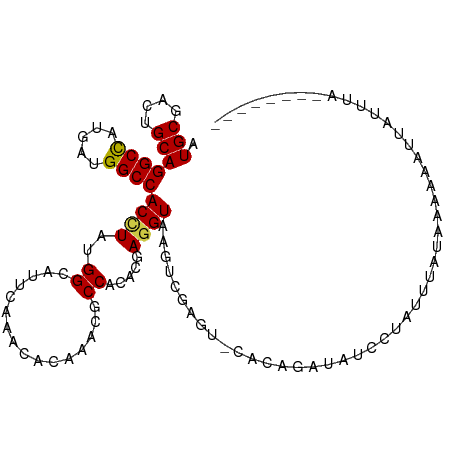
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -10.46 |
| Energy contribution | -10.45 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.984060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
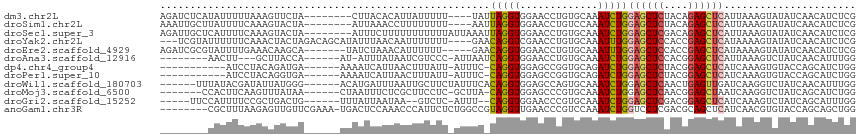
>dm3.chr2L 8193006 104 - 23011544 AGAUCUCAUAUUUUUAAAGUUCUA--------CUUACACAUUAUUUUU----UAUUAGGUGGAACCUGUGCAAAUCUGGAGCUCUACAGAGCUCAUUAAAGUAUAUCAACAUCUCG ((((..............((((((--------((((............----...)))))))))).(((((.......((((((....))))))......))))).....)))).. ( -21.98, z-score = -2.55, R) >droSim1.chr2L 7976052 104 - 22036055 AAAUUGCUUAUUUUCAAAGUACUA--------AUUAAACCUUUUUUUU----AAUUAGGUGGAACCUGUCCAAAUCUGGAGCUCUACAGAGCUCAUUAAAGUAUAUCAACAUCUCG ....(((((............(((--------((((((.......)))----)))))).((((.....))))......((((((....))))))....)))))............. ( -21.00, z-score = -2.61, R) >droSec1.super_3 3689939 108 - 7220098 AGAUUGCUCAUUUUCAAAGUACUA--------AUUUCUUUUUUUUUUUAUUAAAUUAGGUGGAACCUGUGCAAAUCUGGAGCUCGACAGAGCUCAUUAAAGUAUAUCAACAUCUCG (((((((.((.(((((.....(((--------((((...............))))))).)))))..)).)).))))).((((((....))))))...................... ( -21.36, z-score = -1.51, R) >droYak2.chr2L 10825150 109 - 22324452 ---UCGUAUUUUUUCAAACUACUAGACAGCAUAUUUAACAAUUUUUUU----GAACAGGUCGAACCUGUGCAAAUUUGGAGCUCCACCGAGCUCAUAAAAGUAUAUCAACAUCUCG ---..((((((((.........................(((....(((----(.(((((.....))))).)))).)))((((((....))))))..))))))))............ ( -22.80, z-score = -2.87, R) >droEre2.scaffold_4929 17099090 104 - 26641161 AGAUCGCGUAUUUUGAAACAAGCA-------UAUCUAAACAUUUUUU-----GAACAGGUGGAACCUGUGCAAAUUUGGAGCUCCACCGAGCUCAUAAAAGUAUAUCAACAUCUCG .(((...(((((((..........-------........((...(((-----(.(((((.....))))).))))..))((((((....))))))...))))))))))......... ( -23.40, z-score = -1.82, R) >droAna3.scaffold_12916 9537605 97 - 16180835 --------AACUU---GCUUACCA------AU-AUUUAUAAUCGUCCC-AUUAAUCAGGUGGAACCUGUGCAAAUCUGGAGCUCCACGGAGCUCAUUAAAGUCUAUCAACAUUUGG --------.....---.....(((------(.-..........((.((-(((.....))))).)).(((......((.((((((....)))))).....)).......))).)))) ( -18.52, z-score = -0.51, R) >dp4.chr4_group4 1628170 97 - 6586962 -----------AUCCUACAGAUGA------AAAAUCAUUAACUUUAUU-AUUUC-CAGGUGGAGCCGGUGCAGAUCUGGAGCUCUACGGAGCUCAUCAAAGUGUACCAGCAUCUGG -----------......((((((.------..................-..(((-(....))))..((((((......((((((....)))))).......))))))..)))))). ( -32.32, z-score = -3.20, R) >droPer1.super_10 631570 97 - 3432795 -----------AUCCUACAGGUGA------AAAAUCAUUAACUUUAUU-AUUUC-CAGGUGGAGCCGGUGCAGAUCUGGAGCUCUACGGAGCUCAUCAAAGUGUACCAGCAUCUGG -----------......((((((.------..................-..(((-(....))))..((((((......((((((....)))))).......))))))..)))))). ( -31.72, z-score = -2.71, R) >droWil1.scaffold_180703 2863496 104 - 3946847 ------UUUAUACGAUAUUAUGGG------ACAUGAUUUAAUUGCUUCUAUUUCACAGGUGGAGCCAGUGCAAAUCUGGAGCUCAACUGAGUUGAUCAAGGUCUAUCAACAUUUGG ------......(((.....((((------.((.(((((.((((.(((((((.....))))))).))))..)))))))...)))).....((((((........))))))..))). ( -20.60, z-score = 0.31, R) >droMoj3.scaffold_6500 30797726 101 + 32352404 -------CCACUUCAAGUUUAUAA------CUAAUUUCUCGCUUCCUC-GCUUA-CAGGUGGAGCCCGUGCAAAUCUGGAGCUCAACGGAGCUAAUCAAGGUCUAUCAGCAUCUGG -------.................------..........((......-))...-(((((((((((((........))).))))...(((.((.....)).))).....)))))). ( -20.30, z-score = 0.44, R) >droGri2.scaffold_15252 5091621 100 + 17193109 -----UUCCAUUUUCCGCUGACUG------UUUAUUAAUAA--GUCUC-AUUU--CAGGUGGAACCCGUGCAAAUCUGGAGCUCGACGGAGCUCAUCAAAGUCUAUCAGCAUUUGG -----((((((((......((((.------..........)--)))..-....--.))))))))((.((((.......((((((....))))))......(.....).))))..)) ( -22.32, z-score = -0.35, R) >anoGam1.chr3R 30910563 107 - 53272125 --------CGCUUUAAGAGUUGUUCGAAA-UGACUCCAAACCCAUUCUCUGGCCGUAGGUUGAACCCGUCCAAAUCUGGUCCUCGACGCAGCUCAUCAACGUGUACCAGCAGCUGG --------........(((((((((((..-.(((.......(((.....)))...((((((...........))))))))).)))).)))))))...........((((...)))) ( -21.20, z-score = 1.15, R) >consensus ________UAUUUUCAAAGUACUA______AUAUUUAAUAAUUUUUUU_AUUUA_CAGGUGGAACCUGUGCAAAUCUGGAGCUCUACGGAGCUCAUCAAAGUAUAUCAACAUCUGG .......................................................(((((.............)))))((((((....))))))...................... (-10.46 = -10.45 + -0.01)
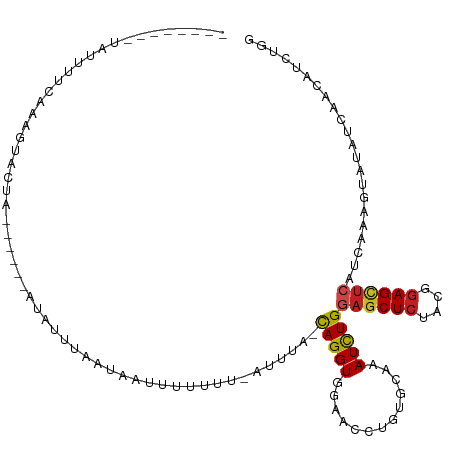
| Location | 8,193,081 – 8,193,181 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.91 |
| Shannon entropy | 0.70652 |
| G+C content | 0.45589 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -10.44 |
| Energy contribution | -10.30 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.53 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 8193081 100 - 23011544 AUGCGACUGCAGGCUAUGAUGGCCACCUAUGGCAUUCAAACACAAACCCCCCAUGAGGUAAGUCCGGU-CACAGAUCUCAUAUUUUUAAAGUUCUACUUAC------- .((.(((((..((((.....))))((((((((..................)))).)))).....))))-).))............................------- ( -18.57, z-score = 0.49, R) >droSim1.chr2L 7976127 100 - 22036055 AUGCGACUGCAGGCUAUGAUGGCCACCUAUGGCAUUCAAACACAAACCCCACACGAGGUAAGUCGAGU-CAUAAAUUGCUUAUUUUCAAAGUACUAAUUAA------- ....((((...((((.(((..((((....))))..))).......(((.(....).))).)))).)))-)......(((((.......)))))........------- ( -17.90, z-score = 0.26, R) >droSec1.super_3 3690018 100 - 7220098 AUGCGACUGCAGGCUAUGAUGGCCACCUAUGGCAUCCAAACACAAACCCCACACGAGGUAAAUCGAGU-CGCAGAUUGCUCAUUUUCAAAGUACUAAUUUC------- ..((((((((.((((.((((.((((....))))............(((.(....).)))..)))))))-))))).))))......................------- ( -22.90, z-score = -1.12, R) >droYak2.chr2L 10825225 97 - 22324452 AUGCGACUGCAGGCUAUGAUGGCCACCUAUGGCAUUCAAACACAGACGCCACAUGAGGUAAGU----U-CGUAUUUUUUCAAACUACUAGACAGCAUAUUUA------ ((((((((...((((.....))))((((.((((..((.......)).))))....)))).)).----)-)))))............................------ ( -19.30, z-score = -0.14, R) >droEre2.scaffold_4929 17099164 101 - 26641161 AUGCGACUGCAGGCUAUGAUGGCCACCUAUGGCAUUCAAACACAGACGCCACACGAGGUAAGUCGAAA-CGUAGAUCGCGUAUUUUGAAACAAGCAUAUCUA------ ((((((((((.((((.....))))((((.((((..((.......)).))))....)))).........-.)))).)))))).....................------ ( -26.50, z-score = -1.26, R) >droAna3.scaffold_12916 9537680 95 - 16180835 AUGCGCCUGCAGGCCAUGAUGGCCACCUACGGCAUUCAAACCCAAACGCCGCACGAGGUGAGUC--AA-UAGUUAUAACUUGCUUACCAAUAUUUAUA---------- ...((..(((.((((.....))))......(((..............)))))))).(((((((.--..-.(((....))).)))))))..........---------- ( -24.04, z-score = -0.87, R) >dp4.chr4_group4 1628244 99 - 6586962 AUGCGACUGCAGGCCAUGAUGGCCACCUACGGCAUUCAAACACAGACGCCACACGAGGUAAGUCGAUUACAAUCUCAUCCUACAGAUGAAAAAUCAUUA--------- ...(((((...((((.....))))((((..(((..((.......)).))).....)))).))))).........(((((.....)))))..........--------- ( -24.70, z-score = -2.19, R) >droPer1.super_10 631644 99 - 3432795 AUGCGACUGCAGGCCAUGAUGGCCACCUACGGCAUUCAAACACAGACGCCACACGAAGUAAGUCGAUUACAAUCUCAUCCUACAGGUGAAAAAUCAUUA--------- ...(((((((.((((.....))))......(((..((.......)).))).......)).))))).........(((((.....)))))..........--------- ( -22.40, z-score = -1.21, R) >droWil1.scaffold_180703 2863571 97 - 3946847 AUGCGUUUGCAGGCCAUGAUGGCCACCUAUGGCAUACAAACUCAAACGCCACAUGAGGUAAGGAUUGCUUUAUACGAUA----UUAUGGGACAUGAUUUAA------- .(((....)))((((.....)))).((((((((..............)))..((((((((.....))))))))......----..)))))...........------- ( -22.44, z-score = -0.28, R) >droMoj3.scaffold_6500 30797795 108 + 32352404 AUGCGCCUGCAGGCCAUGAUGGCCACCUAUGGCAUACAAACGCAAACCCCUCAUGAGGUGAGUGAGUACAGUUCUUCCACUUCAAGUUUAUAACUAAUUUCUCGCUUC .((((..((...((((((..((...))))))))...))..))))..........((((((((.((((..((((.....((.....))....)))).)))))))))))) ( -23.20, z-score = 0.13, R) >droGri2.scaffold_15252 5091695 99 + 17193109 AUGCGUCUGCAGGCCAUGAUGGCCACUUAUGGCAUUCAAACCCAGACACCGCAUGAGGUACAACUUUG-AGCAGUUCCAUUUUCCGCUGACUGUUUAUUA-------- ....(((((..((((.....))))......((........)))))))(((......))).......((-(((((((............)))))))))...-------- ( -24.10, z-score = -0.26, R) >anoGam1.chr3R 30910641 100 - 53272125 AUGCGCCUGCAGGCCAUGAUGGCCACGUACGGCAUUCAAACGCAAACACCACAUGAAGUAAGUA-UGCUCUCCGCUUUAAGAGUUGUUCGAAAUGACUCCA------- ..((((((((.((((.....))))..))).)))........(((.((...((.....))..)).-))).....)).....((((..(.....)..))))..------- ( -24.00, z-score = 0.04, R) >consensus AUGCGACUGCAGGCCAUGAUGGCCACCUAUGGCAUUCAAACACAAACGCCACACGAGGUAAGUCGAGU_CACAGAUAUCCUAUUUAUAAAAAAUUAUUUA________ .(((....)))((((.....))))((((..((................)).....))))................................................. (-10.44 = -10.30 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:29 2011