
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,551,656 – 17,551,755 |
| Length | 99 |
| Max. P | 0.979966 |

| Location | 17,551,656 – 17,551,755 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 74.48 |
| Shannon entropy | 0.51830 |
| G+C content | 0.47110 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.979966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
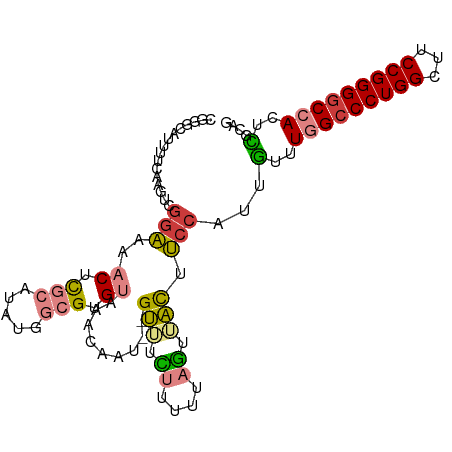
>dm3.chr3R 17551656 99 + 27905053 CGGGCAUUUUCAAGUCGGAAAACUCGCAUAUGGCGUGUAAACAAU--GUUUCUUUUUAGUAACUUCCAUUGUUUGGCCCUGGCUUCCGGGGCCACUCGCAG ((((..(((((......)))))))))......(((.(((((((((--(..................))))))))((((((((...)))))))))).))).. ( -30.67, z-score = -2.19, R) >droPer1.super_3 6760826 82 + 7375914 ---------------CGGGUAUCCCAUA---GUCGAGCUAUCGUCUGGAAUGCCUCCAUGUUUCCCCAUUGUUUAGACCUGGCUUCCGGGGC-ACUUGUGA ---------------(((((..(((..(---(.(((....))).))((((.(((.....((((...........))))..))))))))))..-)))))... ( -22.70, z-score = 0.06, R) >droAna3.scaffold_13340 18121773 98 - 23697760 -UUACAUCUUCAAGUUAUACAUCUUAAAAAUAGCGUGCUAACUUUUUGUAUUUUUUUUGUUACUU--AUUUUUUAGCCCUGGCUUCCGGGGCCAUUGGCAG -............(((((...........))))).((((((......(((..........)))..--........(((((((...)))))))..)))))). ( -18.40, z-score = -0.97, R) >droEre2.scaffold_4770 13633657 99 - 17746568 CGGGUAUUUUCAAGUCGGAAAACUCGCAUAUGGCGUGUAAACAAU--GUUUCUUCGUAGUUACUUCCAUUGUUUGGCCCUGGCUUCCGGGGCCAUUCGCAG .((((.(((((.....)))))))))((..((((...((((...((--(......)))..))))..)))).(..(((((((((...)))))))))..))).. ( -31.80, z-score = -2.69, R) >droYak2.chr3R 6388661 99 + 28832112 UGGGUAUUUUCAAGUCGGAAAACUCGCAUAUGGCGUGUAAACAAU--GUUUCUUAUUAGUUACUUCCAUUGUUUGGCCCUGGCUUCCGGGGCCAUUCGCAG .((((.(((((.....)))))))))((..((((...((((..(((--(.....))))..))))..)))).(..(((((((((...)))))))))..))).. ( -32.20, z-score = -2.89, R) >droSec1.super_6 418169 99 - 4358794 UGGGCAUUUUCGAGUCGGAAAACUCGCAUGUGGCGUGUAAACAAU--GUUUCUUUUUAGUAACUUCCAUUGUUUGGCCCUGGCUUCCGGGGCCACUCGCAG ....(((...(((((......)))))...)))(((.(((((((((--(..................))))))))((((((((...)))))))))).))).. ( -33.37, z-score = -2.30, R) >droSim1.chr3R 23600310 99 - 27517382 CGGGCAUUUUCAAGUCGGAAAACUCGCAUGUGGCGUGUAAACAAU--GUUUCUUUUUAGUAACUUCCAUUGUUUGGCCCUGGCUUCCGGGGCCACUCGCAG ((((..(((((......)))))))))......(((.(((((((((--(..................))))))))((((((((...)))))))))).))).. ( -30.67, z-score = -1.82, R) >consensus CGGGCAUUUUCAAGUCGGAAAACUCGCAUAUGGCGUGUAAACAAU__GUUUCUUUUUAGUUACUUCCAUUGUUUGGCCCUGGCUUCCGGGGCCACUCGCAG ................(((..((.(((.....))).)).........(((.((....)).))).)))...(..(((((((((...)))))))))..).... (-17.12 = -17.60 + 0.48)

| Location | 17,551,656 – 17,551,755 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 74.48 |
| Shannon entropy | 0.51830 |
| G+C content | 0.47110 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -12.92 |
| Energy contribution | -13.74 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17551656 99 - 27905053 CUGCGAGUGGCCCCGGAAGCCAGGGCCAAACAAUGGAAGUUACUAAAAAGAAAC--AUUGUUUACACGCCAUAUGCGAGUUUUCCGACUUGAAAAUGCCCG ..(((.(((((((.((...)).)))))((((((((..................)--))))))))).)))(((...((((((....))))))...))).... ( -27.47, z-score = -1.89, R) >droPer1.super_3 6760826 82 - 7375914 UCACAAGU-GCCCCGGAAGCCAGGUCUAAACAAUGGGGAAACAUGGAGGCAUUCCAGACGAUAGCUCGAC---UAUGGGAUACCCG--------------- ......((-(.((((((((((....(((.....)))(....).....))).))))((.(((....))).)---)..))).)))...--------------- ( -19.70, z-score = 0.42, R) >droAna3.scaffold_13340 18121773 98 + 23697760 CUGCCAAUGGCCCCGGAAGCCAGGGCUAAAAAAU--AAGUAACAAAAAAAAUACAAAAAGUUAGCACGCUAUUUUUAAGAUGUAUAACUUGAAGAUGUAA- .(((...((((((.((...)).))))))......--...((((................))))))).((.(((((((((........)))))))))))..- ( -18.29, z-score = -0.94, R) >droEre2.scaffold_4770 13633657 99 + 17746568 CUGCGAAUGGCCCCGGAAGCCAGGGCCAAACAAUGGAAGUAACUACGAAGAAAC--AUUGUUUACACGCCAUAUGCGAGUUUUCCGACUUGAAAAUACCCG ..(((...(((((.((...)).)))))((((((((..................)--)))))))...)))..(((.((((((....))))))...))).... ( -26.07, z-score = -2.17, R) >droYak2.chr3R 6388661 99 - 28832112 CUGCGAAUGGCCCCGGAAGCCAGGGCCAAACAAUGGAAGUAACUAAUAAGAAAC--AUUGUUUACACGCCAUAUGCGAGUUUUCCGACUUGAAAAUACCCA ..(((...(((((.((...)).)))))((((((((..................)--)))))))...)))..(((.((((((....))))))...))).... ( -26.07, z-score = -2.19, R) >droSec1.super_6 418169 99 + 4358794 CUGCGAGUGGCCCCGGAAGCCAGGGCCAAACAAUGGAAGUUACUAAAAAGAAAC--AUUGUUUACACGCCACAUGCGAGUUUUCCGACUCGAAAAUGCCCA ..(((.(((((((.((...)).)))))((((((((..................)--))))))))).)))..(((.((((((....))))))...))).... ( -30.17, z-score = -2.44, R) >droSim1.chr3R 23600310 99 + 27517382 CUGCGAGUGGCCCCGGAAGCCAGGGCCAAACAAUGGAAGUUACUAAAAAGAAAC--AUUGUUUACACGCCACAUGCGAGUUUUCCGACUUGAAAAUGCCCG ..(((.(((((((.((...)).)))))((((((((..................)--))))))))).)))..(((.((((((....))))))...))).... ( -28.07, z-score = -1.92, R) >consensus CUGCGAGUGGCCCCGGAAGCCAGGGCCAAACAAUGGAAGUAACUAAAAAGAAAC__AUUGUUUACACGCCAUAUGCGAGUUUUCCGACUUGAAAAUGCCCG .......((((((.((...)).)))))).....((((((...........((((.....))))...(((.....)))...))))))............... (-12.92 = -13.74 + 0.82)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:13 2011