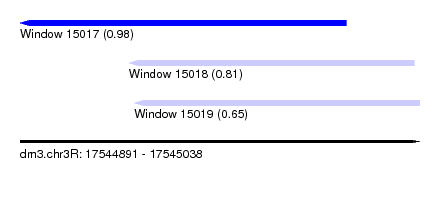
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,544,891 – 17,545,038 |
| Length | 147 |
| Max. P | 0.977767 |

| Location | 17,544,891 – 17,545,011 |
|---|---|
| Length | 120 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 76.75 |
| Shannon entropy | 0.45713 |
| G+C content | 0.55629 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -23.54 |
| Energy contribution | -23.91 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
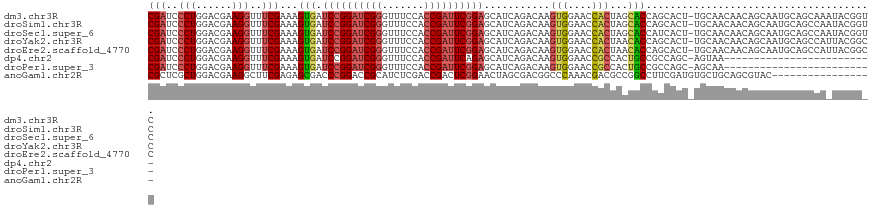
>dm3.chr3R 17544891 120 - 27905053 CGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAGCACCAGCACU-UGCAACAACAGCAAUGCAGCAAAUACGGUC (((..(((......)))..)))...(((.((((((((((.......))))))))))((........((((....)))).......(((.(-(((.......))))))).))...))).... ( -39.30, z-score = -2.62, R) >droSim1.chr3R 23593517 120 + 27517382 CGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAGCACCAGCACU-UGCAACAACAGCAAUGCAGCCAAUACGGUC (((..(((......)))..)))...(((.((((((((((.......))))))))))..........((((....))))..)))..(((.(-(((.......))))))).(((.....))). ( -39.20, z-score = -2.43, R) >droSec1.super_6 411492 120 + 4358794 CGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAGCACCAUCACU-UGCAACAACAGCAAUGCAGCCAAUACGGUC (((..(((......)))..))).((((((((((((((((.......))))))))))..........((((....)))).......)))))-)(((..........))).(((.....))). ( -37.50, z-score = -2.03, R) >droYak2.chr3R 6380746 120 - 28832112 CGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAACACCAGCACU-UGCAACAACAGCAAUGCAGCCAUUACGGCC (((..(((......)))..)))...(((.((((((((((.......))))))))))..........((((....))))..)))..(((.(-(((.......))))))).(((.....))). ( -41.50, z-score = -3.38, R) >droEre2.scaffold_4770 13627274 120 + 17746568 CGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAACACCAGCACU-UGCAACAACAGCAAUGCAGCCAUUACGGCC (((..(((......)))..)))...(((.((((((((((.......))))))))))..........((((....))))..)))..(((.(-(((.......))))))).(((.....))). ( -41.50, z-score = -3.38, R) >dp4.chr2 23974080 95 - 30794189 CGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCAGAGCAUCAGACAAGUGGAACCGCCACUGCCGCCAGC-AGUAA------------------------- (((..(((......)))..)))...(((.((.(((((((.......))))))).)).))).......(((....))).(((((.....))-)))..------------------------- ( -28.20, z-score = -0.55, R) >droPer1.super_3 6752125 95 - 7375914 CGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCGCCACUGCCGCCAGC-AGCAA------------------------- (((..(((......)))..)))...(((.((((((((((.......)))))))))).)))......(((((.....)))))((.(....)-.))..------------------------- ( -33.80, z-score = -1.52, R) >anoGam1.chr2R 45106435 104 + 62725911 CGCUCGCUGGACGAAGGCUUCGAGAGCGACCCGGACCGCAUCUCGACCGACUCGGAACUAGCGACGGCCCAAACGACGCCGGCCUUCGAUGUGCUGCAGCGUAC----------------- (((((((....(((((((((((((((((........))).))))))(((...))).....(((.((.......)).))).))))))))....)).).))))...----------------- ( -37.40, z-score = -0.24, R) >consensus CGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUACCACCAGCACU_UGCAACAACAGCAAUGCAGCCA_UACGG_C (((..(((......)))..)))...(((.((((((((((.......))))))))))...........(((....)))...)))...................................... (-23.54 = -23.91 + 0.37)
| Location | 17,544,931 – 17,545,036 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 87.03 |
| Shannon entropy | 0.29747 |
| G+C content | 0.53100 |
| Mean single sequence MFE | -34.91 |
| Consensus MFE | -23.75 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
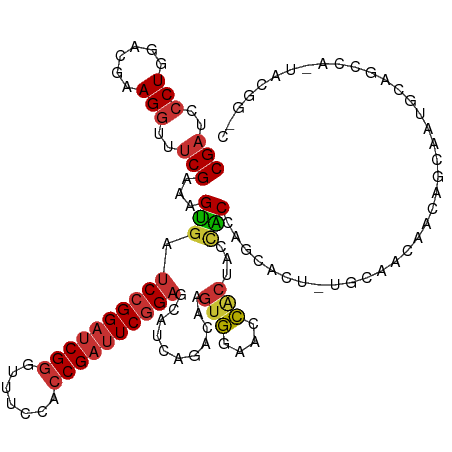
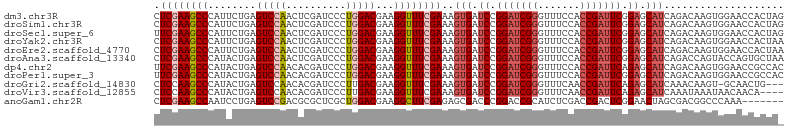
>dm3.chr3R 17544931 105 - 27905053 CUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAG .((((((((..(((...(((((..........))))))))))))))))..(((.((((((((((.......)))))))))).)))......((((....)))).. ( -38.80, z-score = -2.48, R) >droSim1.chr3R 23593557 105 + 27517382 CUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAG .((((((((..(((...(((((..........))))))))))))))))..(((.((((((((((.......)))))))))).)))......((((....)))).. ( -38.80, z-score = -2.48, R) >droSec1.super_6 411532 105 + 4358794 UUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAG (((((((((..(((...(((((..........))))))))))))))))).(((.((((((((((.......)))))))))).)))......((((....)))).. ( -39.60, z-score = -2.84, R) >droYak2.chr3R 6380786 105 - 28832112 CUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAA .((((((((..(((...(((((..........))))))))))))))))..(((.((((((((((.......)))))))))).)))......((((....)))).. ( -38.80, z-score = -2.46, R) >droEre2.scaffold_4770 13627314 105 + 17746568 CUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUAA .((((((((..(((...(((((..........))))))))))))))))..(((.((((((((((.......)))))))))).)))......((((....)))).. ( -38.80, z-score = -2.46, R) >droAna3.scaffold_13340 18115228 105 + 23697760 CUCGAAGCCCAUACUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACCAGUACCAGUGCUAA .....((((..(((((.(((....((((..(((......)))..))))..(((.((((((((((.......)))))))))).)))..))))))))...).))).. ( -38.80, z-score = -2.92, R) >dp4.chr2 23974095 105 - 30794189 UUCGAAGCCCAUACUGAGUCCAACACGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCAGAGCAUCAGACAAGUGGAACCGCCAC (((((((((........(((((..........)))))...))))))))).(((.((.(((((((.......))))))).)).))).......((((.....)))) ( -32.00, z-score = -1.34, R) >droPer1.super_3 6752140 105 - 7375914 UUCGAAGCCCAUACUGAGUCCAACACGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCGCCAC (((((((((........(((((..........)))))...))))))))).(((.((((((((((.......)))))))))).))).......((((.....)))) ( -38.10, z-score = -2.57, R) >droGri2.scaffold_14830 4850696 101 + 6267026 CUCCAAGCCCAUACUGAGUCCAACACGAUCCCUUGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCAACCGAUUCAGAGCAUCAAACAAGU-GCAACUG--- .............(((((((.....((((((...(((....))).(....)......))))))(((....)))))))))).((((.......))-)).....--- ( -26.10, z-score = -1.26, R) >droVir3.scaffold_12855 1251747 101 + 10161210 CUCCAAGCCCAUACUGAGUCCAACACGAUCCCUUGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCAACCGAUUCAGAGCAUCAAAUAAAUAACAACA---- .............(((((((.....((((((...(((....))).(....)......))))))(((....)))))))))).....................---- ( -23.00, z-score = -1.19, R) >anoGam1.chr2R 45106466 98 + 62725911 CUCGAAGCCAAUCCUGAGUCCGACGCGCUCGCUGGACGAAGGCUUCGAGAGCGACCCGGACCGCAUCUCGACCGACUCGGAACUAGCGACGGCCCAAA------- ......(((......((((.......))))(((((.(((.((..(((((((((........))).))))))))...)))...)))))...))).....------- ( -31.20, z-score = 0.30, R) >consensus CUCGAAGCCCAUACUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACUA_ .((((((((........(((((..........)))))...))))))))..(((.((.(((((((.......))))))).)).))).................... (-23.75 = -24.50 + 0.75)
| Location | 17,544,933 – 17,545,038 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 86.84 |
| Shannon entropy | 0.30010 |
| G+C content | 0.53558 |
| Mean single sequence MFE | -34.41 |
| Consensus MFE | -23.75 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
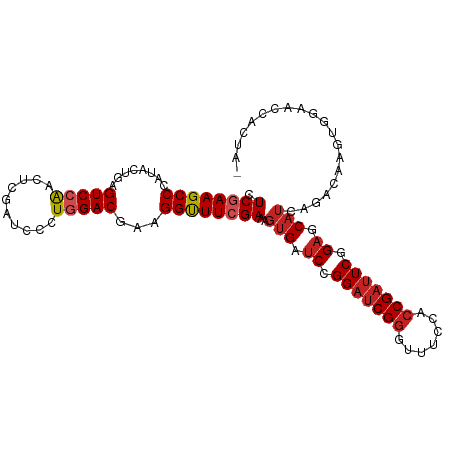
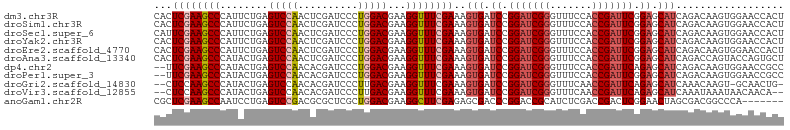
>dm3.chr3R 17544933 105 - 27905053 CACUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACU (((((((((((..(((...(((((..........))))))))))))))))..))).((((((((((.......))))))))))..........((((....)))) ( -38.10, z-score = -2.14, R) >droSim1.chr3R 23593559 105 + 27517382 CACUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACU (((((((((((..(((...(((((..........))))))))))))))))..))).((((((((((.......))))))))))..........((((....)))) ( -38.10, z-score = -2.14, R) >droSec1.super_6 411534 105 + 4358794 CAUUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACU ..(((((((((..(((...(((((..........))))))))))))))))).(((.((((((((((.......)))))))))).)))......((((....)))) ( -39.20, z-score = -2.57, R) >droYak2.chr3R 6380788 105 - 28832112 CACUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACU (((((((((((..(((...(((((..........))))))))))))))))..))).((((((((((.......))))))))))..........((((....)))) ( -38.10, z-score = -2.14, R) >droEre2.scaffold_4770 13627316 105 + 17746568 CACUCGAAGCCCAUUCUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACU (((((((((((..(((...(((((..........))))))))))))))))..))).((((((((((.......))))))))))..........((((....)))) ( -38.10, z-score = -2.14, R) >droAna3.scaffold_13340 18115230 105 + 23697760 CACUCGAAGCCCAUACUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACCAGUACCAGUGCU .......((((..(((((.(((....((((..(((......)))..))))..(((.((((((((((.......)))))))))).)))..))))))))...).))) ( -38.10, z-score = -2.59, R) >dp4.chr2 23974097 103 - 30794189 --UUCGAAGCCCAUACUGAGUCCAACACGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCAGAGCAUCAGACAAGUGGAACCGCC --(((((((((........(((((..........)))))...))))))))).(((.((.(((((((.......))))))).)).))).......(((....))). ( -30.70, z-score = -0.96, R) >droPer1.super_3 6752142 103 - 7375914 --UUCGAAGCCCAUACUGAGUCCAACACGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCGCC --(((((((((........(((((..........)))))...))))))))).(((.((((((((((.......)))))))))).))).......(((....))). ( -36.80, z-score = -2.20, R) >droGri2.scaffold_14830 4850696 101 + 6267026 --CUCCAAGCCCAUACUGAGUCCAACACGAUCCCUUGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCAACCGAUUCAGAGCAUCAAACAAGU-GCAACUG- --.............(((((((.....((((((...(((....))).(....)......))))))(((....)))))))))).((((.......))-)).....- ( -26.10, z-score = -1.26, R) >droVir3.scaffold_12855 1251747 101 + 10161210 --CUCCAAGCCCAUACUGAGUCCAACACGAUCCCUUGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCAACCGAUUCAGAGCAUCAAAUAAAUAACAACA-- --.............(((((((.....((((((...(((....))).(....)......))))))(((....)))))))))).....................-- ( -23.00, z-score = -1.19, R) >anoGam1.chr2R 45106468 98 + 62725911 CGCUCGAAGCCAAUCCUGAGUCCGACGCGCUCGCUGGACGAAGGCUUCGAGAGCGACCCGGACCGCAUCUCGACCGACUCGGAACUAGCGACGGCCCA------- .((.((..((.....(((((((.(..((..(((.....)))..)).(((((((((........))).))))))).))))))).....))..))))...------- ( -32.20, z-score = 0.67, R) >consensus CACUCGAAGCCCAUACUGAGUCCAACUCGAUCCCUGGACGAAGGUUUCGAAAGUGAUCCGGAUCGGGUUUCCACCGAUUCGGAGCAUCAGACAAGUGGAACCACU ...((((((((........(((((..........)))))...))))))))..(((.((.(((((((.......))))))).)).))).................. (-23.75 = -24.50 + 0.75)
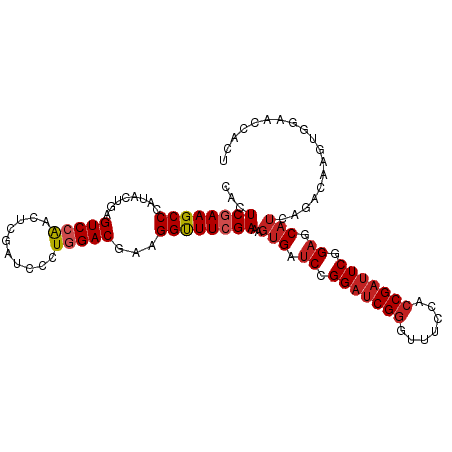
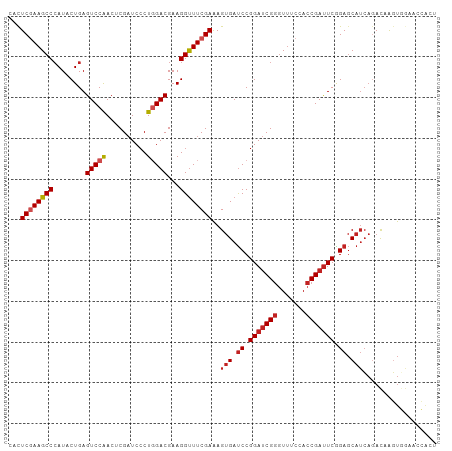
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:09 2011