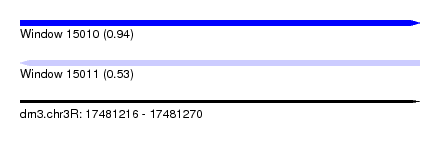
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,481,216 – 17,481,270 |
| Length | 54 |
| Max. P | 0.940204 |

| Location | 17,481,216 – 17,481,270 |
|---|---|
| Length | 54 |
| Sequences | 8 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.54987 |
| G+C content | 0.43016 |
| Mean single sequence MFE | -8.84 |
| Consensus MFE | -5.69 |
| Energy contribution | -5.67 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
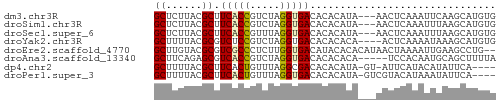
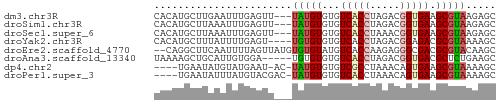
>dm3.chr3R 17481216 54 + 27905053 GCUCUUACGCUUCACCGUCUAGGUGACACACAUA---AACUCAAAUUCAAGCAUGUG ((......)).(((((.....)))))..(((((.---..((........)).))))) ( -8.70, z-score = -1.31, R) >droSim1.chr3R 23528094 54 - 27517382 GCUCUUACGCUUCACCGUCUAGGUGACACACAUA---AACUCAAAUUUAAGCAUGUG ((......)).(((((.....)))))..(((((.---..((........)).))))) ( -8.70, z-score = -1.27, R) >droSec1.super_6 347212 54 - 4358794 GCUCUUACGCUUCACCGUUUAGGUGACACACAUA---AACUCAAAUUUAAGCAUGUG ((......)).(((((.....)))))..(((((.---..((........)).))))) ( -8.70, z-score = -1.21, R) >droYak2.chr3R 6319360 53 + 28832112 GCUUUUACGCGUCUCCGUCUAGGUGACACACACA----ACUCAAAAUAAAGCAUGUG ((......))(((.((.....)).)))...((((----.((........))..)))) ( -8.60, z-score = -0.64, R) >droEre2.scaffold_4770 13562088 55 - 17746568 GCUUGUACGCGUCGCCCUCUUGGUGACAUACACACAUAACUAAAAUUGAAGCCUG-- (((((....)((((((.....)))))).....................))))...-- ( -10.50, z-score = -1.26, R) >droAna3.scaffold_13340 18051938 52 - 23697760 GCUUCAGAGCGUCACCGUCUAGGUGACACACACA-----UCCACAAUGCAGCUUUUA .....(((((((((((.....)))))).....((-----(.....)))..))))).. ( -14.10, z-score = -2.79, R) >dp4.chr2 23895687 51 + 30794189 GCUUUUACGCUUCACUGUUUAGGCGACACACAUA-GU-AUUCAUACAUAUUCA---- ((((..(((......)))..))))..........-((-(....))).......---- ( -5.20, z-score = -1.01, R) >droPer1.super_3 6669703 52 + 7375914 GCUUUUACGCUUCACUGUUUAGGUGACACACAUA-GUCGUACAUAAAUAUUCA---- .....(((((((((((.....))))).......)-).))))............---- ( -6.21, z-score = -0.37, R) >consensus GCUUUUACGCUUCACCGUCUAGGUGACACACAUA___AACUCAAAUUUAAGCAUGUG ((......)).(((((.....)))))............................... ( -5.69 = -5.67 + -0.02)
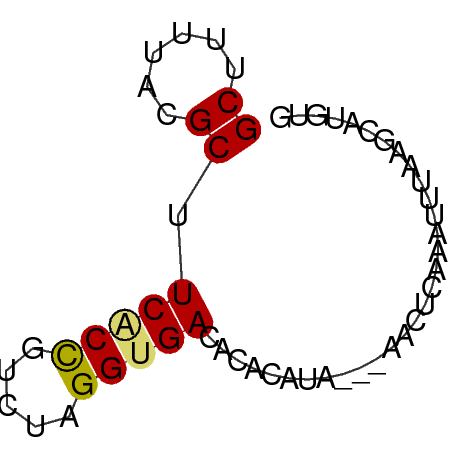
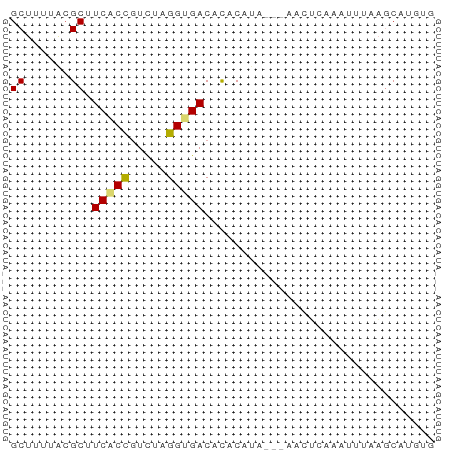
| Location | 17,481,216 – 17,481,270 |
|---|---|
| Length | 54 |
| Sequences | 8 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.54987 |
| G+C content | 0.43016 |
| Mean single sequence MFE | -11.89 |
| Consensus MFE | -5.97 |
| Energy contribution | -6.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
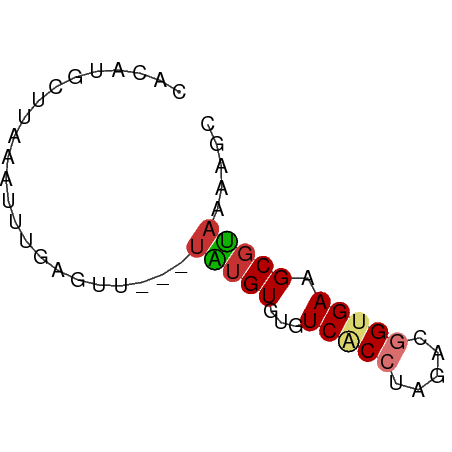
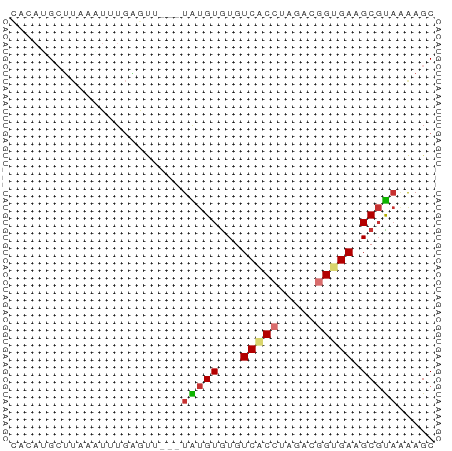
>dm3.chr3R 17481216 54 - 27905053 CACAUGCUUGAAUUUGAGUU---UAUGUGUGUCACCUAGACGGUGAAGCGUAAGAGC .....((((......))))(---(((((...(((((.....))))).)))))).... ( -12.90, z-score = -1.18, R) >droSim1.chr3R 23528094 54 + 27517382 CACAUGCUUAAAUUUGAGUU---UAUGUGUGUCACCUAGACGGUGAAGCGUAAGAGC .....(((((....)))))(---(((((...(((((.....))))).)))))).... ( -13.30, z-score = -1.56, R) >droSec1.super_6 347212 54 + 4358794 CACAUGCUUAAAUUUGAGUU---UAUGUGUGUCACCUAAACGGUGAAGCGUAAGAGC .....(((((....)))))(---(((((...(((((.....))))).)))))).... ( -13.30, z-score = -1.77, R) >droYak2.chr3R 6319360 53 - 28832112 CACAUGCUUUAUUUUGAGU----UGUGUGUGUCACCUAGACGGAGACGCGUAAAAGC ((((.((((......))))----)))).(((((.((.....)).)))))........ ( -14.00, z-score = -1.43, R) >droEre2.scaffold_4770 13562088 55 + 17746568 --CAGGCUUCAAUUUUAGUUAUGUGUGUAUGUCACCAAGAGGGCGACGCGUACAAGC --..((((........))))...((((..((((.((.....)).))))..))))... ( -12.00, z-score = -0.48, R) >droAna3.scaffold_13340 18051938 52 + 23697760 UAAAAGCUGCAUUGUGGA-----UGUGUGUGUCACCUAGACGGUGACGCUCUGAAGC .....((((((((...))-----)))(.((((((((.....)))))))).)...))) ( -16.20, z-score = -2.09, R) >dp4.chr2 23895687 51 - 30794189 ----UGAAUAUGUAUGAAU-AC-UAUGUGUGUCGCCUAAACAGUGAAGCGUAAAAGC ----...............-..-(((((...((((.......)))).)))))..... ( -6.50, z-score = -0.04, R) >droPer1.super_3 6669703 52 - 7375914 ----UGAAUAUUUAUGUACGAC-UAUGUGUGUCACCUAAACAGUGAAGCGUAAAAGC ----..................-(((((...((((.......)))).)))))..... ( -6.90, z-score = 0.16, R) >consensus CACAUGCUUAAAUUUGAGUU___UAUGUGUGUCACCUAGACGGUGAAGCGUAAAAGC .......................(((((...(((((.....))))).)))))..... ( -5.97 = -6.30 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:03 2011