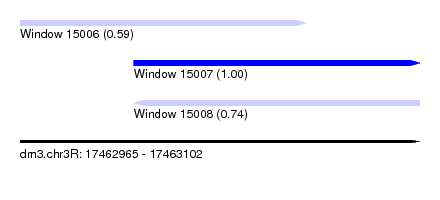
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,462,965 – 17,463,102 |
| Length | 137 |
| Max. P | 0.996321 |

| Location | 17,462,965 – 17,463,063 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.74 |
| Shannon entropy | 0.39376 |
| G+C content | 0.50640 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
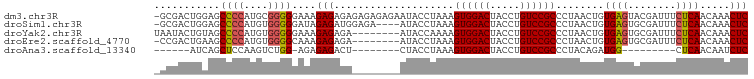
>dm3.chr3R 17462965 98 + 27905053 -GCGACUGGAGCCCCAUGCGGGGGAAAGAGAGAGAGAGAGAAUACCUAAAGUGGACUACCUGUCCGCCCUAACUGUGAGUACGAUUUCUCAACAAACUC -.......(((((((....))))....((((((..(.....(((..((..((((((.....))))))..))..))).....)..))))))......))) ( -24.70, z-score = -0.34, R) >droSim1.chr3R 23510280 94 - 27517382 -GCGACUGGAGCCCCAUGUGGGGGAUAGAGAUGGAGA----AUACCUAAAGUGGACUACCUGUCCGCCCUAACUGUGAGUGCGAUUUCUCAACAAACUC -....(((...((((.....)))).))).....(((.----.........((((((.....))))))......((((((........))).)))..))) ( -23.30, z-score = 0.13, R) >droYak2.chr3R 6300636 91 + 28832112 UAAUACUGUAGCCCCAUGUGGGGGAAAGAGAGA--------AUACCAAAAGUGGACUACCUGUCCGCCCUAACUGUGAGUGCGAUUUCUCAACAAACUC ......(((..((((....))))....((((((--------.........((((((.....))))))......(((....))).)))))).)))..... ( -23.50, z-score = -1.36, R) >droEre2.scaffold_4770 13544063 90 - 17746568 -CCGACUGAAGCCCCAUGUGGGGCAAAGAGAGA--------AUACCUAAAGUGGACUACCUGUCCGCCCUAACUGUGAGUGCGAUUUCUCAACAAACUC -.........(((((....)))))...((((((--------.........((((((.....))))))......(((....))).))))))......... ( -26.70, z-score = -2.59, R) >droAna3.scaffold_13340 18035900 75 - 23697760 ------AUCAGCUCCAAGUCUGG-AGAGAGACU--------CUACCUAAAGUGGACUACCUGUCCGCCCUACAGAUGG---------CUCAACAAUCUC ------.....(((((....)))-)).(((..(--------((.......((((((.....)))))).....)))...---------)))......... ( -18.90, z-score = -0.63, R) >consensus _GCGACUGGAGCCCCAUGUGGGGGAAAGAGAGA________AUACCUAAAGUGGACUACCUGUCCGCCCUAACUGUGAGUGCGAUUUCUCAACAAACUC ...........((((....))))....(((....................((((((.....))))))........((((........)))).....))) (-17.84 = -18.12 + 0.28)
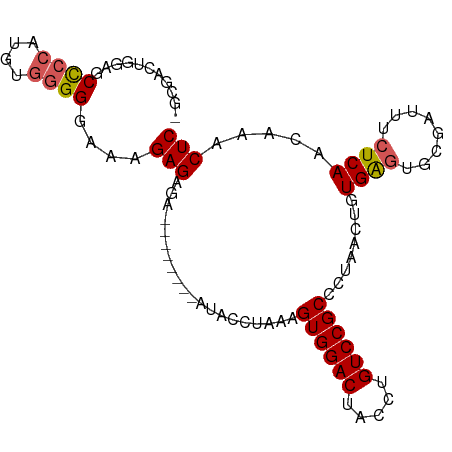
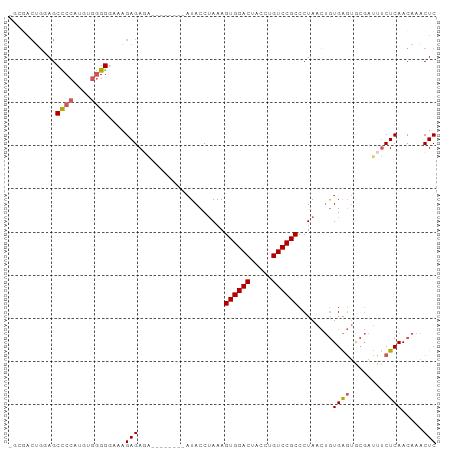
| Location | 17,463,004 – 17,463,102 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.07 |
| Shannon entropy | 0.48760 |
| G+C content | 0.47074 |
| Mean single sequence MFE | -20.99 |
| Consensus MFE | -12.00 |
| Energy contribution | -13.56 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.996321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17463004 98 + 27905053 AAUACCUAAAGUGGACU----ACCUGUCCGCCCUAACUGUGAGUACGAUUUCUCAACAAACUCACAGAUACUCUGCAAACGCAUACAACAAACUACCGGACU ..........((((((.----....))))))((...((((((((...............))))))))......(((....)))..............))... ( -22.76, z-score = -3.92, R) >droSim1.chr3R 23510315 97 - 27517382 AAUACCUAAAGUGGACU----ACCUGUCCGCCCUAACUGUGAGUGCGAUUUCUCAACAAACUCACAGAUACUCUGCAAACGCACACAGCGAACUACCGACU- ..........((((((.----....)))))).....((((((((...............))))))))............(((.....)))...........- ( -23.76, z-score = -3.42, R) >droYak2.chr3R 6300668 97 + 28832112 AAUACCAAAAGUGGACU----ACCUGUCCGCCCUAACUGUGAGUGCGAUUUCUCAACAAACUCACAGAUACUCUGCAAACGCACAGAGCGACCAACCGACU- ..........((((((.----....)))))).....((((((((...............))))))))...(((((........))))).............- ( -25.56, z-score = -3.70, R) >droEre2.scaffold_4770 13544094 98 - 17746568 AAUACCUAAAGUGGACU----ACCUGUCCGCCCUAACUGUGAGUGCGAUUUCUCAACAAACUCACAGAUACUCUGCAAGCGCACACAGCGACCAACCGACUU ..........((((((.----....)))))).....(((((.((((((....))..........(((.....)))...)))).))))).............. ( -23.50, z-score = -2.64, R) >droAna3.scaffold_13340 18035925 85 - 23697760 UCUACCUAAAGUGGACU----ACCUGUCCGCCCUACAGAUGG---------CUCAACAAUCUCACAGAUACUAU--AACCGCUCACCAACAGAUACUUCU-- (((.......((((((.----....))))))......(((((---------.......(((.....))).....--..))).))......))).......-- ( -12.34, z-score = -0.85, R) >droPer1.super_3 6650354 92 + 7375914 CACAGUAAAACUGGACUGCCUACCUGUCCGCCCUAACUGCGAGUACGAUUACUCAACAG-CACACACAGAC-----AGAUACUUUCUGAGACAGACUU---- ..(((.....)))..........(((((........(((.(((((....)))))..)))-..........(-----(((.....)))).)))))....---- ( -18.00, z-score = -1.25, R) >consensus AAUACCUAAAGUGGACU____ACCUGUCCGCCCUAACUGUGAGUGCGAUUUCUCAACAAACUCACAGAUACUCUGCAAACGCACACAGCGACCUACCGACU_ ..........((((((.........)))))).....((((((((...............))))))))................................... (-12.00 = -13.56 + 1.56)

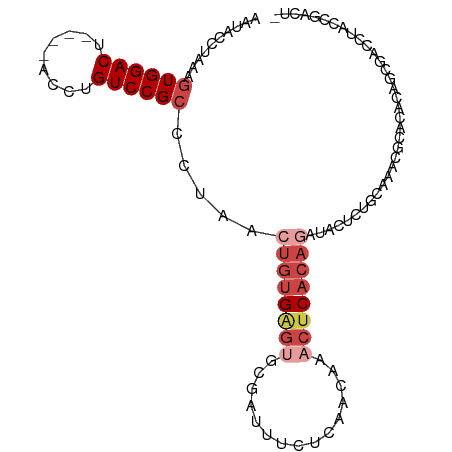
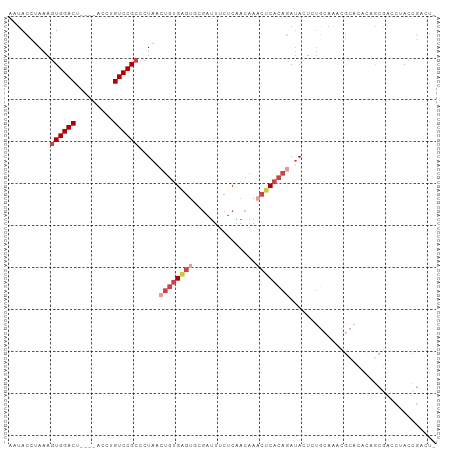
| Location | 17,463,004 – 17,463,102 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 74.07 |
| Shannon entropy | 0.48760 |
| G+C content | 0.47074 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -12.97 |
| Energy contribution | -15.08 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17463004 98 - 27905053 AGUCCGGUAGUUUGUUGUAUGCGUUUGCAGAGUAUCUGUGAGUUUGUUGAGAAAUCGUACUCACAGUUAGGGCGGACAGGU----AGUCCACUUUAGGUAUU .((((((((.(((((...........))))).))))(((((((....(((....))).)))))))....))))((((....----.))))............ ( -30.60, z-score = -2.98, R) >droSim1.chr3R 23510315 97 + 27517382 -AGUCGGUAGUUCGCUGUGUGCGUUUGCAGAGUAUCUGUGAGUUUGUUGAGAAAUCGCACUCACAGUUAGGGCGGACAGGU----AGUCCACUUUAGGUAUU -(.((.((((..(((.....))).)))).)).)..((((((((..((.((....))))))))))))((((((.((((....----.)))).))))))..... ( -29.30, z-score = -1.55, R) >droYak2.chr3R 6300668 97 - 28832112 -AGUCGGUUGGUCGCUCUGUGCGUUUGCAGAGUAUCUGUGAGUUUGUUGAGAAAUCGCACUCACAGUUAGGGCGGACAGGU----AGUCCACUUUUGGUAUU -..((((..(((((((((((......)))))))..((((((((..((.((....))))))))))))....)))((((....----.)))).)..)))).... ( -31.50, z-score = -1.89, R) >droEre2.scaffold_4770 13544094 98 + 17746568 AAGUCGGUUGGUCGCUGUGUGCGCUUGCAGAGUAUCUGUGAGUUUGUUGAGAAAUCGCACUCACAGUUAGGGCGGACAGGU----AGUCCACUUUAGGUAUU .............((((((((((((..(((.....)))..))).....((....))))))..)))))(((((.((((....----.)))).)))))...... ( -30.50, z-score = -1.18, R) >droAna3.scaffold_13340 18035925 85 + 23697760 --AGAAGUAUCUGUUGGUGAGCGGUU--AUAGUAUCUGUGAGAUUGUUGAG---------CCAUCUGUAGGGCGGACAGGU----AGUCCACUUUAGGUAGA --.....((((((..((((..(((((--.....(((.....))).....))---------))((((((.......))))))----.)..)))).)))))).. ( -21.20, z-score = -0.30, R) >droPer1.super_3 6650354 92 - 7375914 ----AAGUCUGUCUCAGAAAGUAUCU-----GUCUGUGUGUG-CUGUUGAGUAAUCGUACUCGCAGUUAGGGCGGACAGGUAGGCAGUCCAGUUUUACUGUG ----..(.((((((........((((-----((((((.(..(-((((.(((((....))))))))))..).)))))))))))))))).)(((.....))).. ( -32.60, z-score = -3.25, R) >consensus _AGUCGGUAGGUCGCUGUGUGCGUUUGCAGAGUAUCUGUGAGUUUGUUGAGAAAUCGCACUCACAGUUAGGGCGGACAGGU____AGUCCACUUUAGGUAUU ...................................((((((((....(((....))).))))))))((((((.((((.........)))).))))))..... (-12.97 = -15.08 + 2.11)
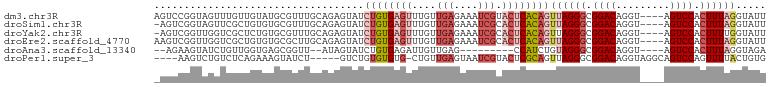
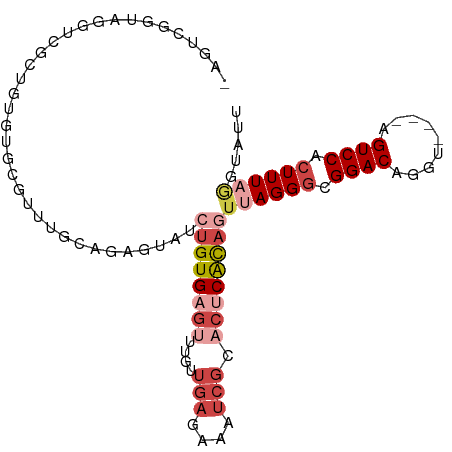
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:33:00 2011