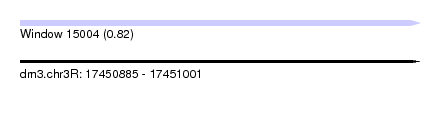
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,450,885 – 17,451,001 |
| Length | 116 |
| Max. P | 0.820642 |

| Location | 17,450,885 – 17,451,001 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.01 |
| Shannon entropy | 0.62017 |
| G+C content | 0.51812 |
| Mean single sequence MFE | -35.99 |
| Consensus MFE | -11.29 |
| Energy contribution | -10.86 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.81 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
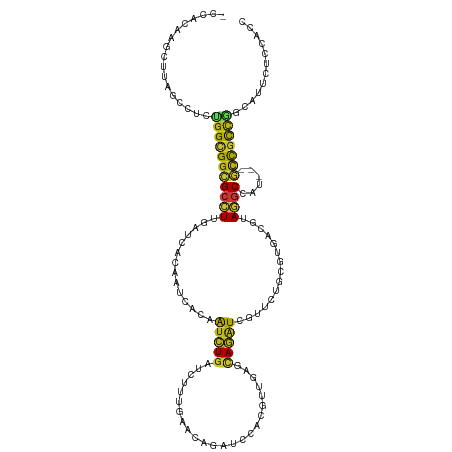
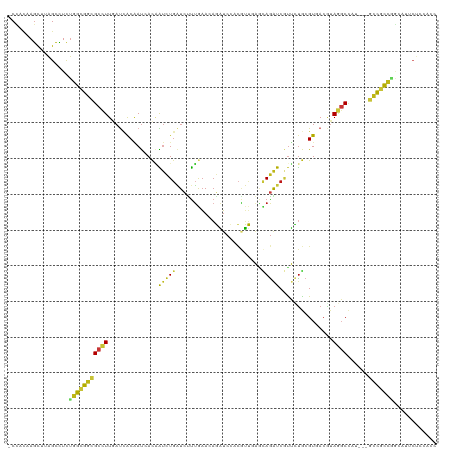
>dm3.chr3R 17450885 116 + 27905053 -CCACAAGCUUAGCCUCUGGCGGCGCCUUGAUCACGAUAACGAUCUGAUCUUUGAAGAGAUCCACGUUGAGCAGGUCGUUCUGCGUGACGUAGGCCAU---GCCGCCGGAGUUCUCCACC -.............((((((((((((((((.(((((..(((((((((.((..((..(....)..))..)).)))))))))...))))))).))))...---))))))))))......... ( -47.60, z-score = -3.55, R) >droGri2.scaffold_14906 2515690 116 - 14172833 -CCACUAGUUUGGCUUCUGGCGGCGCCUUGAUCACAAUCACAAUCUGAUCCUUGAACAGAUCCACAUUGAGCAGAUCAUUCUGCGUCACAUAGGCCAU---GCCACCGCUGUUCUCCACC -.....((..((((...(((((..((((((((..((((....(((((.((...)).)))))....)))).(((((....)))))))))...))))..)---))))..))))..))..... ( -30.80, z-score = -1.28, R) >droMoj3.scaffold_6540 1742512 116 + 34148556 -CUACAAGUUUGGCUUCUGGUGGCGCUUUGAUCACGAUCACAAUCUGAUCUUUAAAGAGAUCCACGUUGAGCAGAUCGCUCUGCGUUACGUACGCCAU---GCCGCCCACAUUCUCCACC -......(..((((.....((((((((((((....(((((.....))))).))))))......((((...(((((....)))))...)))).))))))---))))..)............ ( -31.70, z-score = -1.51, R) >droVir3.scaffold_13047 12886711 116 - 19223366 -CCACAAGUUUGGCCUCUGGCGGCGCCUUGAUCACAAUCACAAUCUGAUCUUUGAACAGAUCCACGUUGAGCAGAUCGUUUUGCGUCACGUAGGCCAU---GCCGCCAAUAUUCUCCACC -(((......)))....(((((((((((((((....))))......(((((......))))).((((.(((((((....))))).))))))))))...---)))))))............ ( -32.40, z-score = -1.41, R) >droWil1.scaffold_181089 3892944 116 - 12369635 -CCACAAGCUUGGCCUCUGGCGGCGCCUUAAUCACAAUAACAAUCUGAUCUUUAAACAGAUCAACAUUCAAUAGAUCAUUUUGCGUUACAUAGGCCAA---ACCGCCGGCAUUCUCCACU -......(((((((((.((..(((((...................((((((......))))))....((....)).......))))).)).)))))))---...)).((......))... ( -24.00, z-score = -1.51, R) >droSim1.chr3R 23497290 116 - 27517382 -CCACAAGCUUAGCCUCUGGCGGCGCCUUGAUCACGAUGACGAUCUGAUCUUUGAAGAGAUCCACGUUGAGCAGGUCGUUCUGCGUGACGUAGGCCAU---GCCGCCGGAGUUCUCCACC -.............((((((((((((((((.(((((..(((((((((.((..((..(....)..))..)).)))))))))...))))))).))))...---))))))))))......... ( -48.00, z-score = -3.25, R) >droSec1.super_6 317429 116 - 4358794 -CCACAAGCUUAGCCUCUGGCGGCGCCUUGAUCACGAUGACGAUCUGAUCUUUGAAGAGAUCCACGUUGAGCAGGUCGUUCUGCGUGACGUAGGCCAU---GCCGCCGGAGUUCUCCACC -.............((((((((((((((((.(((((..(((((((((.((..((..(....)..))..)).)))))))))...))))))).))))...---))))))))))......... ( -48.00, z-score = -3.25, R) >droYak2.chr3R 6288313 116 + 28832112 -CCACAAGCUUAGCCUCUGGCGGCGCCUUGAUCACUAUUACGAUCUGAUCUUUGAAGAGAUCCACGUUGAGCAGGUCGUUCUGCGUGACGUAGGCCAU---GCCGCCGGAGUUCUCCACC -.............((((((((((((((.((((........)))).((((((....)))))).(((((..(((((....)))))..)))))))))...---))))))))))......... ( -45.10, z-score = -3.14, R) >droEre2.scaffold_4770 13531884 116 - 17746568 -CCACAAGCUUAGCCUCUGGCGGCGCCUUGAUCACGAUAACGAUCUGAUCUUUGAAGAGAUCCACGUUGAGCAGGUCGUUCUGCGUGACGUAGGCCAU---GCCGCCGGAGUUCUCCACC -.............((((((((((((((((.(((((..(((((((((.((..((..(....)..))..)).)))))))))...))))))).))))...---))))))))))......... ( -47.60, z-score = -3.55, R) >droAna3.scaffold_13340 18024917 119 - 23697760 -CCACUAGUUUGGCCUCUGGCGGCGCCUUAAUCACAAUCACUAUCUGAUCUUUAAACAGAUCCACAUUGAGAAGAUCAUUUUGCGUCACAUAGGCCAUCAUGCCGCCAGCAUUCUCCACC -.........(((...((((((((((((...((.((((....(((((.........)))))....)))).)).(((........)))....))))......))))))))......))).. ( -26.90, z-score = -1.29, R) >apiMel3.Group9 4685532 106 - 10282195 -CAUGAAGUGUAGCUUCUGGUGGAGCUUUCACCGCCAUUAUAGCUUGGUCUUUGUACAUUGGGACAGAACGUAAAUCAUGAUACGUUACAUAAGCGUAUUGUCUGUC------------- -.(..(((...((((..((((((........))))))....))))....)))..).......((((((..(.....)..((((((((.....)))))))).))))))------------- ( -27.70, z-score = -0.88, R) >dp4.chr2 23860829 116 + 30794189 -CCACAAGCUUGGCCUCUGGUGGCGCCUUGAUUACAAUCACAAUCUGAUCUUUGAACAGAUCCACAUUCAAUAGAUCAUUUUGCGUCACAUAGGCCAU---GCCGCCGGCAUUCUCCACU -.........((((((...(((((((..((((....)))).....(((((((((((..........))))).))))))....)))))))..))))))(---(((...))))......... ( -34.20, z-score = -2.93, R) >droPer1.super_3 6637472 116 + 7375914 -CCACAAGCUUGGCCUCUGGUGGCGCCUUGAUUACAAUCACAAUCUGAUCUUUGAACAGAUCCACAUUCAAUAGAUCAUUUUGCGUCACAUAGGCCAU---GCCGCCGGCAUUCUCCACU -.........((((((...(((((((..((((....)))).....(((((((((((..........))))).))))))....)))))))..))))))(---(((...))))......... ( -34.20, z-score = -2.93, R) >anoGam1.chr2R 49685318 113 - 62725911 -CCUCCAGUUUAGCCUCUGGUGGUGCCUUAAUAACAACGAUCGUCUGCUCGCGGAACAUCUCCAGCGAGCUCAGAUCCUGACAGGUAAAGUAGCCUUGCUUCGUGUGCGCCAUC------ -...((((........))))((((((............((((....(((((((((.....))).))))))...))))...(((.(..(((((....)))))).)))))))))..------ ( -35.60, z-score = -1.62, R) >triCas2.chrUn_90 81353 110 - 150287 CCGAUAAAUGC-GUUUUCGGGGGCGCUUUGACCACGAUGACCGUGUUCUGCUUGAACUUCUCUAUCGACCUCAGGUCCUGAUACGUGACAAACCCGUGCUUGUCGUUGUUC--------- .((((....((-((..((((((.(((...((.((((.....)))).)).)).(((...((......))..)))).)))))).))))(((((........)))))))))...--------- ( -26.10, z-score = 0.61, R) >consensus _CCACAAGCUUAGCCUCUGGCGGCGCCUUGAUCACAAUCACAAUCUGAUCUUUGAACAGAUCCACGUUGAGCAGAUCGUUCUGCGUGACGUAGGCCAU___GCCGCCGGCAUUCUCCACC .................(((((((((((..............(((((........................)))))...............))))......)))))))............ (-11.29 = -10.86 + -0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:57 2011