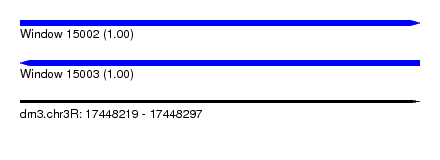
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,448,219 – 17,448,297 |
| Length | 78 |
| Max. P | 0.999791 |

| Location | 17,448,219 – 17,448,297 |
|---|---|
| Length | 78 |
| Sequences | 14 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Shannon entropy | 0.53559 |
| G+C content | 0.47718 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -17.05 |
| Energy contribution | -16.27 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.997521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
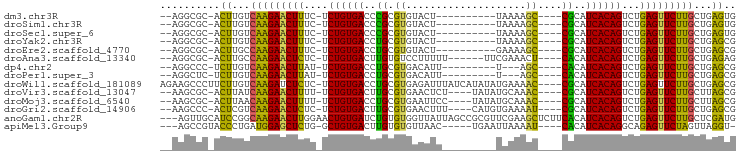
>dm3.chr3R 17448219 78 + 27905053 CACUCAGCAAGAACUCAGACUGUGAUGCG----GCUUUUA----------AGUACACGCGGGUCACAGA-GAAAGUUCUUGACAAGU-GCGCCU-- ((((...((((((((....(((((((.((----(((....----------))).....)).))))))).-...))))))))...)))-).....-- ( -25.40, z-score = -1.82, R) >droSim1.chr3R 23494731 78 - 27517382 CACUCAGCAAGAACUCAGACUGUGAUGCG----GCUUUUA----------AGUACACGCGGGUCACAGA-GAAAGUUCUUGACAAGU-GCGCCU-- ((((...((((((((....(((((((.((----(((....----------))).....)).))))))).-...))))))))...)))-).....-- ( -25.40, z-score = -1.82, R) >droSec1.super_6 314853 78 - 4358794 CACUCAGCAAGAACUCAGACUGUGAUGCG----GCUUUUA----------AGUACACGCGGGUCACAGA-GAAAGUUCUUGACAAGU-GCGCCU-- ((((...((((((((....(((((((.((----(((....----------))).....)).))))))).-...))))))))...)))-).....-- ( -25.40, z-score = -1.82, R) >droYak2.chr3R 6285774 78 + 28832112 CGCUCAGCAAGAACUCAGACUGUGAUGCG----GCUUUUA----------AGUACACGCAGGUCACAGA-GAAAGUUCUUGACAAGU-GCGCCU-- ((((...((((((((....((((((((((----.......----------......))))..)))))).-...))))))))...)))-).....-- ( -25.12, z-score = -1.49, R) >droEre2.scaffold_4770 13529313 78 - 17746568 CGCUCAGCAAGAACUCAGACUGUGAUGCG----GCUUUUC----------AGUACACGCAGGUCACAGA-GAAAGUUCUUGGCAAGU-GCGCCU-- .((....((((((((....((((((((((----.......----------......))))..)))))).-...))))))))((....-))))..-- ( -24.52, z-score = -0.67, R) >droAna3.scaffold_13340 18021997 82 - 23697760 CUCUCAGCAAGAACUCAGACUGUGAUGUG----AGUUUCGAA------AAAAAGGACACAAGUCACAGA-GAGAGUUCUUGGCAAGU-GCGCCU-- .......(((((((((...(((((((.((----.((((....------.....)))).)).))))))).-..)))))))))((....-))....-- ( -26.00, z-score = -1.68, R) >dp4.chr2 23857562 76 + 30794189 CGCUCAGCAAGAACUCAGACUGUGAUGUG----GCU---A---------AAUGUCACGCAGGUCACAGA-AUAAGUUCUUGACAAGA-GGGCCU-- .((((..((((((((.(..((((((((((----((.---.---------...)))))....))))))).-.).))))))))......-))))..-- ( -27.90, z-score = -3.08, R) >droPer1.super_3 6634125 76 + 7375914 CGCUCAGCAAGAACUCAGACUGUGAUGUG----GCU---A---------AAUGUCACGCAGGUCACAGA-AUAAGUUCUUGACAAGA-GAGCCU-- .((((..((((((((.(..((((((((((----((.---.---------...)))))....))))))).-.).))))))))......-))))..-- ( -28.80, z-score = -3.47, R) >droWil1.scaffold_181089 3890514 91 - 12369635 CGCUCAGCAAGAACUCAGACUGUGAUGCG----GUUUUCAUAUAUGAUAAAUCUCACGCAGGUCACAGA-GAGAGAUCUUGACAAGAAGGGCUUCU .((((..(((((.(((...((((((((((----(((((((....))).))))....))))..)))))).-..))).))))).......)))).... ( -24.00, z-score = -0.76, R) >droVir3.scaffold_13047 12884336 84 - 19223366 CGCUAAGCAAGAACUCAGACUGUGAUGCG----GUUUGCAUAUA----AGAGUUCACGCAAGUCACAGA-AAAAGUUCUUGAUAAGU-GCGCUU-- ((((...((((((((....((((((((((----....((.....----...))...))))..)))))).-...))))))))...)))-).....-- ( -26.30, z-score = -2.35, R) >droMoj3.scaffold_6540 1740124 84 + 34148556 CGCUAAGCAAGAACUCAGACUGUGAUGCG----GUUUGCAUAUA----GGAAUUCACGCAGGUCACAGA-AAAAGUUCUUGUUAAGU-GCGCUU-- ((((.((((((((((....((((((((((----..((.(.....----).))....))))..)))))).-...)))))))))).)))-).....-- ( -28.40, z-score = -2.97, R) >droGri2.scaffold_14906 2513267 84 - 14172833 CGCUCAGCAAGAACUCAGACUGUGAUGCG----AUUUUCACAUG----AAAGUUCACGCAAGUCACAGA-GAGAGUUCUUGACGAGU-GGGCUU-- (((((..(((((((((...((((((((((----((((((....)----)))))...))))..)))))).-..)))))))))..))))-).....-- ( -36.00, z-score = -4.46, R) >anoGam1.chr2R 49683801 93 - 62725911 CAUCGAGCAAGAACUCAGACUGUGAUGUGAAGAGCUUCGAACGCGGCUAAUAACCACACAGAUCACAGUUCCAAGUUCUUGCCGGAUGCAACU--- ((((..(((((((((..(((((((((.((....((.......))((.......))...)).)))))))))...)))))))))..)))).....--- ( -33.80, z-score = -4.22, R) >apiMel3.Group9 4684073 82 - 10282195 -ACCUAACUAGAACUCUGCCUGUGAUGUG----AUUUUAAUUCA-----GUUAACACACAAGUCACAGC-CAGAGCUCCAUCAGGGUACGGCU--- -((((.....((.(((((.((((((((((----...(((((...-----)))))..))))..)))))).-))))).)).....))))......--- ( -23.00, z-score = -2.05, R) >consensus CGCUCAGCAAGAACUCAGACUGUGAUGCG____GCUUUCA_________AAGUACACGCAGGUCACAGA_GAAAGUUCUUGACAAGU_GCGCCU__ ..((...((((((((....((((((((((...........................))))..)))))).....))))))))...)).......... (-17.05 = -16.27 + -0.79)

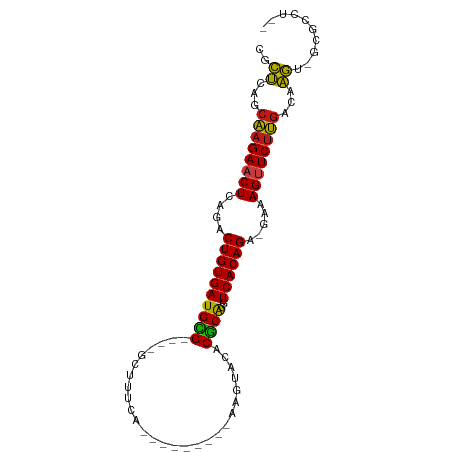
| Location | 17,448,219 – 17,448,297 |
|---|---|
| Length | 78 |
| Sequences | 14 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 74.22 |
| Shannon entropy | 0.53559 |
| G+C content | 0.47718 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -18.54 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17448219 78 - 27905053 --AGGCGC-ACUUGUCAAGAACUUUC-UCUGUGACCCGCGUGUACU----------UAAAAGC----CGCAUCACAGUCUGAGUUCUUGCUGAGUG --.....(-(((((.(((((((((..-.((((((...(((.((...----------.....))----))).))))))...))))))))).)))))) ( -29.40, z-score = -3.56, R) >droSim1.chr3R 23494731 78 + 27517382 --AGGCGC-ACUUGUCAAGAACUUUC-UCUGUGACCCGCGUGUACU----------UAAAAGC----CGCAUCACAGUCUGAGUUCUUGCUGAGUG --.....(-(((((.(((((((((..-.((((((...(((.((...----------.....))----))).))))))...))))))))).)))))) ( -29.40, z-score = -3.56, R) >droSec1.super_6 314853 78 + 4358794 --AGGCGC-ACUUGUCAAGAACUUUC-UCUGUGACCCGCGUGUACU----------UAAAAGC----CGCAUCACAGUCUGAGUUCUUGCUGAGUG --.....(-(((((.(((((((((..-.((((((...(((.((...----------.....))----))).))))))...))))))))).)))))) ( -29.40, z-score = -3.56, R) >droYak2.chr3R 6285774 78 - 28832112 --AGGCGC-ACUUGUCAAGAACUUUC-UCUGUGACCUGCGUGUACU----------UAAAAGC----CGCAUCACAGUCUGAGUUCUUGCUGAGCG --...(((-.(..(.(((((((((..-.((((((..((((.((...----------.....))----))))))))))...)))))))))).).))) ( -28.60, z-score = -2.95, R) >droEre2.scaffold_4770 13529313 78 + 17746568 --AGGCGC-ACUUGCCAAGAACUUUC-UCUGUGACCUGCGUGUACU----------GAAAAGC----CGCAUCACAGUCUGAGUUCUUGCUGAGCG --...(((-.(..((.((((((((..-.((((((..((((.((...----------.....))----))))))))))...)))))))))).).))) ( -28.60, z-score = -2.52, R) >droAna3.scaffold_13340 18021997 82 + 23697760 --AGGCGC-ACUUGCCAAGAACUCUC-UCUGUGACUUGUGUCCUUUUU------UUCGAAACU----CACAUCACAGUCUGAGUUCUUGCUGAGAG --......-.(((..(((((((((..-.((((((..((((...((((.------...))))..----))))))))))...)))))))))..))).. ( -27.00, z-score = -2.61, R) >dp4.chr2 23857562 76 - 30794189 --AGGCCC-UCUUGUCAAGAACUUAU-UCUGUGACCUGCGUGACAUU---------U---AGC----CACAUCACAGUCUGAGUUCUUGCUGAGCG --.....(-((..(.((((((((((.-.((((((..((.((......---------.---.))----))..))))))..))))))))))).))).. ( -25.80, z-score = -3.07, R) >droPer1.super_3 6634125 76 - 7375914 --AGGCUC-UCUUGUCAAGAACUUAU-UCUGUGACCUGCGUGACAUU---------U---AGC----CACAUCACAGUCUGAGUUCUUGCUGAGCG --..((((-....(.((((((((((.-.((((((..((.((......---------.---.))----))..))))))..))))))))))).)))). ( -29.50, z-score = -3.96, R) >droWil1.scaffold_181089 3890514 91 + 12369635 AGAAGCCCUUCUUGUCAAGAUCUCUC-UCUGUGACCUGCGUGAGAUUUAUCAUAUAUGAAAAC----CGCAUCACAGUCUGAGUUCUUGCUGAGCG ..........((((.(((((.(((..-.((((((..((((.(...(((((.....)))))..)----))))))))))...))).))))).)))).. ( -23.10, z-score = -1.29, R) >droVir3.scaffold_13047 12884336 84 + 19223366 --AAGCGC-ACUUAUCAAGAACUUUU-UCUGUGACUUGCGUGAACUCU----UAUAUGCAAAC----CGCAUCACAGUCUGAGUUCUUGCUUAGCG --...(((-......(((((((((..-.((((((.(((((((......----..)))))))..----....))))))...)))))))))....))) ( -24.40, z-score = -2.77, R) >droMoj3.scaffold_6540 1740124 84 - 34148556 --AAGCGC-ACUUAACAAGAACUUUU-UCUGUGACCUGCGUGAAUUCC----UAUAUGCAAAC----CGCAUCACAGUCUGAGUUCUUGCUUAGCG --...(((-...((((((((((((..-.((((((..((((........----...........----))))))))))...))))))))).)))))) ( -24.41, z-score = -3.19, R) >droGri2.scaffold_14906 2513267 84 + 14172833 --AAGCCC-ACUCGUCAAGAACUCUC-UCUGUGACUUGCGUGAACUUU----CAUGUGAAAAU----CGCAUCACAGUCUGAGUUCUUGCUGAGCG --......-.((((.(((((((((..-.((((((..((((.....(((----(....))))..----))))))))))...))))))))).)))).. ( -31.90, z-score = -4.51, R) >anoGam1.chr2R 49683801 93 + 62725911 ---AGUUGCAUCCGGCAAGAACUUGGAACUGUGAUCUGUGUGGUUAUUAGCCGCGUUCGAAGCUCUUCACAUCACAGUCUGAGUUCUUGCUCGAUG ---.....((((.((((((((((..((.(((((((....(((((.....)))))....(((....)))..)))))))))..)))))))))).)))) ( -41.80, z-score = -5.59, R) >apiMel3.Group9 4684073 82 + 10282195 ---AGCCGUACCCUGAUGGAGCUCUG-GCUGUGACUUGUGUGUUAAC-----UGAAUUAAAAU----CACAUCACAGGCAGAGUUCUAGUUAGGU- ---........(((((((((((((((-.((((((..((((..((((.-----....))))...----)))))))))).)))))))))).))))).- ( -35.20, z-score = -4.92, R) >consensus __AGGCGC_ACUUGUCAAGAACUUUC_UCUGUGACCUGCGUGAACUU_________UAAAAGC____CGCAUCACAGUCUGAGUUCUUGCUGAGCG ..........((...(((((((((....((((((...(((...........................))).))))))...)))))))))...)).. (-18.54 = -17.72 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:56 2011