| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,427,110 – 17,427,209 |
| Length | 99 |
| Max. P | 0.727060 |

| Location | 17,427,110 – 17,427,209 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 63.23 |
| Shannon entropy | 0.68532 |
| G+C content | 0.37604 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -4.57 |
| Energy contribution | -4.91 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
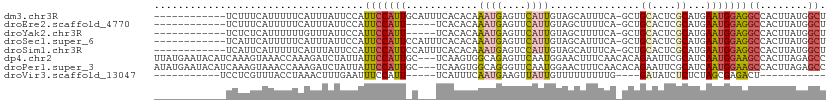
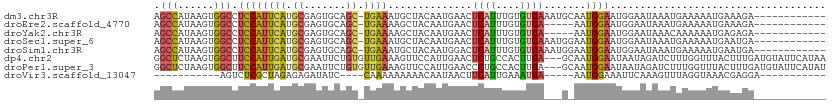
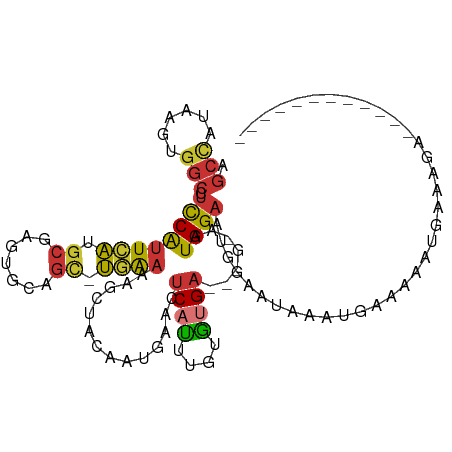
>dm3.chr3R 17427110 99 + 27905053 ------------UCUUUCAUUUUUCAUUUAUUCCAUUCCAUUGCAUUUCACACAAAUGAGUUCAUUGUAGCAUUUCA-GCUGCACUCGCAUGAAUGGAGGCCACUUAUGGCU ------------..................((((((((...(((..........((((....))))(((((......-)))))....))).))))))))((((....)))). ( -26.00, z-score = -3.38, R) >droEre2.scaffold_4770 13511109 94 - 17746568 ------------UCUUUCAUUUUUCAUUUAUUCCAUUCCAUU-----UCACACAAAUGAGUUCAUUGUAGCUUUUCA-GCUGCACUCGCAUGAAUGGAGGCCACUUAUGGCU ------------..................((((((((((((-----(.....))))).((....(((((((....)-))))))...))..))))))))((((....)))). ( -23.00, z-score = -3.07, R) >droYak2.chr3R 6265225 94 + 28832112 ------------UCUCUCAUUUUUUGUUUAUUCCAUUCCAUU-----UCACACAAAUGAGUUCAUUGUAGCUUUUCA-GCUGCACUCGCAUGAAUGGAGGCCACUUAUGGCU ------------..................((((((((((((-----(.....))))).((....(((((((....)-))))))...))..))))))))((((....)))). ( -23.00, z-score = -2.51, R) >droSec1.super_6 296190 99 - 4358794 ------------UCAUUCAUUUUUCAUUUAUUCCAUUCCAUUCCAUUUCACACAAAUGAGUUCAUUGUAGCAUUUCA-GCUGCACUCGCAUGAAUGGAGGCCACUUAUGGCU ------------.......................(((((((((((((.....))))).((....((((((......-))))))...))..))))))))((((....)))). ( -22.50, z-score = -2.68, R) >droSim1.chr3R 23476208 99 - 27517382 ------------UCAUUCAUUUUUCAUUUAUUCCAUUCCAUUCCAUUUCACACAAAUGAGUCCAUUGUAGCAUUUCA-GCUGCACUCGCAUGAAUGGAGGCCACUUAUGGCU ------------.......................(((((((((((((.....))))).((....((((((......-))))))...))..))))))))((((....)))). ( -22.50, z-score = -2.57, R) >dp4.chr2 23812737 109 + 30794189 UUAUGAAUACAUCAAAGUAAACCAAAGAUCUAUUAUUCCAUUGC---UCAAGUGGCAGAGUUCAAUGGAACUUUCAACACAGAAUUCGCAUCAAUGGAAGCCACUUAGAGCC .....((((.(((.............))).))))........((---((((((((((((((((....)))))))..........(((.((....)))))))))))).)))). ( -24.12, z-score = -2.29, R) >droPer1.super_3 6587949 109 + 7375914 AUAUGAAUACAUCAAAGUAAACCAAAGAUCUAUUAUUCCAUUGC---UCAAGUGGCAGGGUUCAAUGGAACUUUCAACACAGAAUUCGCAUCAAUGGAAGCCACUUAGAGCC .....((((.(((.............))).))))........((---((((((((((((((((....)))))))..........(((.((....)))))))))))).)))). ( -23.32, z-score = -1.55, R) >droVir3.scaffold_13047 6380309 81 + 19223366 -----------UCCUCGUUUACCUAAACUUUGAAUUUCCAUU-----UCAUUUCAAUGAAGUUAUUGUUUUUUUUUG----GAUAUCUCUCUAGCGAGACU----------- -----------..((((((((.(((((....((((....(((-----((((....)))))))....))))...))))----).)).......))))))...----------- ( -11.31, z-score = -0.79, R) >consensus ____________UCAUUCAUUUUUCAUUUAUUCCAUUCCAUU_C___UCACACAAAUGAGUUCAUUGUAGCUUUUCA_GCUGCACUCGCAUGAAUGGAGGCCACUUAUGGCU ...................................(((((((............((((....))))...............((....))...)))))))((........)). ( -4.57 = -4.91 + 0.35)
| Location | 17,427,110 – 17,427,209 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 63.23 |
| Shannon entropy | 0.68532 |
| G+C content | 0.37604 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -7.09 |
| Energy contribution | -6.68 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
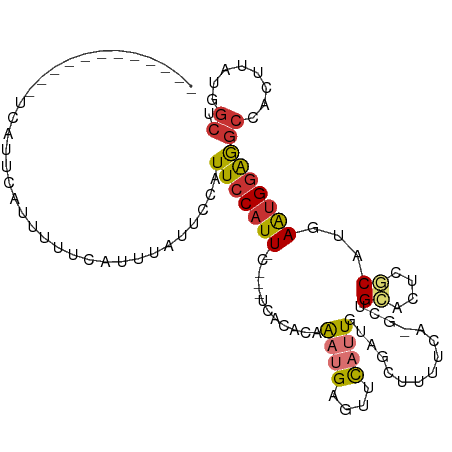
>dm3.chr3R 17427110 99 - 27905053 AGCCAUAAGUGGCCUCCAUUCAUGCGAGUGCAGC-UGAAAUGCUACAAUGAACUCAUUUGUGUGAAAUGCAAUGGAAUGGAAUAAAUGAAAAAUGAAAGA------------ .((((....)))).(((((((.((((..((.(((-......))).))......((((....))))..))))...)))))))...................------------ ( -23.10, z-score = -1.41, R) >droEre2.scaffold_4770 13511109 94 + 17746568 AGCCAUAAGUGGCCUCCAUUCAUGCGAGUGCAGC-UGAAAAGCUACAAUGAACUCAUUUGUGUGA-----AAUGGAAUGGAAUAAAUGAAAAAUGAAAGA------------ .((((....)))).((((((((((((((((.(((-(....))))..........)))))))))).-----))))))........................------------ ( -24.70, z-score = -2.77, R) >droYak2.chr3R 6265225 94 - 28832112 AGCCAUAAGUGGCCUCCAUUCAUGCGAGUGCAGC-UGAAAAGCUACAAUGAACUCAUUUGUGUGA-----AAUGGAAUGGAAUAAACAAAAAAUGAGAGA------------ .((((....)))).((((((((((((((((.(((-(....))))..........)))))))))).-----))))))........................------------ ( -24.70, z-score = -2.52, R) >droSec1.super_6 296190 99 + 4358794 AGCCAUAAGUGGCCUCCAUUCAUGCGAGUGCAGC-UGAAAUGCUACAAUGAACUCAUUUGUGUGAAAUGGAAUGGAAUGGAAUAAAUGAAAAAUGAAUGA------------ .((((....)))).((((((((((((((((.(((-......)))..........)))))))))).)))))).............................------------ ( -21.90, z-score = -1.11, R) >droSim1.chr3R 23476208 99 + 27517382 AGCCAUAAGUGGCCUCCAUUCAUGCGAGUGCAGC-UGAAAUGCUACAAUGGACUCAUUUGUGUGAAAUGGAAUGGAAUGGAAUAAAUGAAAAAUGAAUGA------------ .((((....)))).((((((((((((((((.(((-(.............)).)))))))))))).)))))).............................------------ ( -22.42, z-score = -0.97, R) >dp4.chr2 23812737 109 - 30794189 GGCUCUAAGUGGCUUCCAUUGAUGCGAAUUCUGUGUUGAAAGUUCCAUUGAACUCUGCCACUUGA---GCAAUGGAAUAAUAGAUCUUUGGUUUACUUUGAUGUAUUCAUAA .((((.(((((((.....(..(((.((((((......))..)))))))..).....)))))))))---))....(((((....(((...((....))..))).))))).... ( -29.00, z-score = -2.30, R) >droPer1.super_3 6587949 109 - 7375914 GGCUCUAAGUGGCUUCCAUUGAUGCGAAUUCUGUGUUGAAAGUUCCAUUGAACCCUGCCACUUGA---GCAAUGGAAUAAUAGAUCUUUGGUUUACUUUGAUGUAUUCAUAU .((((.(((((((.....(..(((.((((((......))..)))))))..).....)))))))))---))....(((((....(((...((....))..))).))))).... ( -29.00, z-score = -2.25, R) >droVir3.scaffold_13047 6380309 81 - 19223366 -----------AGUCUCGCUAGAGAGAUAUC----CAAAAAAAAACAAUAACUUCAUUGAAAUGA-----AAUGGAAAUUCAAAGUUUAGGUAAACGAGGA----------- -----------.(((((......))))).((----(................((((((.......-----))))))........((((....))))..)))----------- ( -12.00, z-score = -0.68, R) >consensus AGCCAUAAGUGGCCUCCAUUCAUGCGAGUGCAGC_UGAAAAGCUACAAUGAACUCAUUUGUGUGA___G_AAUGGAAUGGAAUAAAUGAAAAAUGAAAGA____________ .(((......))).((((((((.((.......)).))))..............((((....)))).......)))).................................... ( -7.09 = -6.68 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:52 2011