
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,177,537 – 8,177,647 |
| Length | 110 |
| Max. P | 0.500000 |

| Location | 8,177,537 – 8,177,647 |
|---|---|
| Length | 110 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.59 |
| Shannon entropy | 0.55045 |
| G+C content | 0.40388 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -9.08 |
| Energy contribution | -8.39 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 8177537 110 + 23011544 ----------CAAUCUUCGUCCAAGGCGUGGGUUAAGAUCAUGUACGGCUUCUGGCUGUUUCUGAUCUUCUAUGCCAUGUCCAUACUUAUAUUGAUCUACACAUAUCAAUUCGACAAGUU ----------........(((...((((((((...((((((...(((((.....)))))...))))))))))))))..............((((((........))))))..)))..... ( -29.40, z-score = -2.76, R) >droSim1.chr2L 7960232 110 + 22036055 ----------CAAUCGUCGUCCAAGGCGUGGGUUAAGAUCAUGUACGGCUUCUGGCUGUUUCUGAUCUUCUAUGCCAUGUCCAUACUUAUAUUGAUCUACACAUAUCAAUUCGACAAGUU ----------.....((((..((.((((((((...((((((...(((((.....)))))...)))))))))))))).))..)........((((((........))))))..)))..... ( -32.40, z-score = -3.40, R) >droSec1.super_3 3674594 110 + 7220098 ----------CAAUCGUCGUCCAAGGCGUGGGUUAAGAUCAUGUACGGCUUCUGGCUGUUUCUGAUCUUCUAUGCCAUGUCCAUACUUAUAUUGAUUUACACAUAUCAAUUCGACAAGUU ----------.....((((..((.((((((((...((((((...(((((.....)))))...)))))))))))))).))..)........((((((........))))))..)))..... ( -33.20, z-score = -3.75, R) >droYak2.chr2L 10809552 110 + 22324452 ----------CAAUCGUCGUCCAAGGCAUGGGUUAAGAUCAUGUACGGCUUCUGGCUGUUUCUGAUCUUCUAUGCCAUGUCCAUACUUAUCUUGAUCUACACAUAUCAAUUCGACAAGUU ----------.....((((..((.((((((((...((((((...(((((.....)))))...)))))))))))))).))..).........(((((........)))))...)))..... ( -32.10, z-score = -3.59, R) >droEre2.scaffold_4929 17083559 110 + 26641161 ----------CAAUCGUCGUCCAAGGCGUGGGUUAAGAUCAUGUACGGCUUCUGGCUGUUUCUGAUCUUCUAUGCCAUGUCCAUACUUAUAUUGAUCUACACAUAUCAAUUCGACAAGUU ----------.....((((..((.((((((((...((((((...(((((.....)))))...)))))))))))))).))..)........((((((........))))))..)))..... ( -32.40, z-score = -3.40, R) >droAna3.scaffold_12916 9522588 110 + 16180835 ----------CAAUCAUCGUCCAAGGCCUGGGUGAAGAUCAUGUACGGCUUCUGGCUGUUCCUGAUCUUCUACGCCAUGUCCAUCCUAAUAUUGAUCUACACAUAUCAGUUCGAUAAGUU ----------.....((((..(..(((.(((((((((((((.(.(((((.....))))).).)))))))).)).))).)))...........((((........)))))..))))..... ( -30.70, z-score = -2.27, R) >dp4.chr4_group4 1612104 110 + 6586962 ----------CAGUCCUCGUCCAAGGCCUGGGUGAAGAUUAUGUACGGCUUCUGGCUGUUCCUCAUUUUCUACGCCAUGUCCAUACUGAUUCUGAUCUACACAUAUCAAUUCGACAAGUU ----------(((.(((......))).)))((((.(((..(((.(((((.....)))))....)))..))).)))).((((.....((((..((.....))...))))....)))).... ( -26.60, z-score = -1.01, R) >droPer1.super_10 615421 110 + 3432795 ----------CAGUCCUCGUCCAAGGCAUGGGUGAAGAUCAUGUACGGCUUCUGGCUGUUCCUCAUUUUCUACGCCAUGUCCAUACUGAUUCUGAUCUACACAUAUCAAUUCGACAAGUU ----------........(((...(((((((((((((((.....(((((.....))))).....)))))).)).)))))))......((...((((........))))..)))))..... ( -26.20, z-score = -0.75, R) >droWil1.scaffold_180703 2841314 110 + 3946847 ----------CAAUCAUCGUCCAAGGCAUGGGUUAAGAUUAUGUAUGGAUUUUGGUUAUUCCUGAUAUUCUAUGCCAUGUCCAUAUUGAUAUUGGUCUAUACAUAUCAAUUUGAUAAGUU ----------.(((((..(((((..((((((.......)))))).)))))..))))).....((((((..((.((((((((......)))).)))).))...))))))............ ( -22.70, z-score = 0.16, R) >droVir3.scaffold_12963 9685900 110 + 20206255 ----------CAAUCAUCGUCCAAGGCCUGGGUUAAGAUCAUGUACGGCUUUUGGCUGUUCCUAAUCUUUUACGCCAUGUCCAUAUUGAUAUUGAUCUAUACAUAUCAGUUUGAUAAGUU ----------((((.((((.....(((.(((((.(((((.....(((((.....))))).....)))))..)).))).))).....))))))))......((.((((.....)))).)). ( -22.90, z-score = -0.47, R) >droMoj3.scaffold_6500 30779498 110 - 32352404 ----------CAAUCGUCGUCCAAGGCAUGGGUCAAGAUCAUGUACGGCUUCUGGCUGUUCCUCAUCUUCUAUGCCAUGUCCAUCCUGAUAUUGAUCUAUACAUAUCAAUUCGACAAGUU ----------.....((((..((.((((((((...((((.....(((((.....))))).....)))))))))))).))..).....((.((((((........)))))))))))..... ( -29.20, z-score = -2.28, R) >droGri2.scaffold_15252 5073794 110 - 17193109 ----------CAAUCAUCGUCCAAAGCGUGGGUCAAGAUCAUGUACGGGUUCUGGCUGUUCCUCAUCUUCUAUGCCAUGUCCAUAUUGAUCUUGAUCUAUACAUAUCAAUUUGAUAAGUU ----------...............(..(((((((((((((.(((.(((...((((.................))))..))).))))))))))))))))..).((((.....)))).... ( -28.73, z-score = -2.33, R) >anoGam1.chr3R 22390390 120 - 53272125 UUUCUUUUAACAGUUUUCAUUCAGUGUGUGGCGGAGAGUAAUGUUUACGUUCUGGAUUACCGUCAUCCUGUACUCGAUGGCCAUCUUGAUCUUGGUGUAUCUGCAUCAGUUCGAGCAGUU ........(((.....(((((.((((.(((((((.((..((((....)))).....)).)))))))....)))).)))))....(((((..(((((((....))))))).)))))..))) ( -24.20, z-score = 1.09, R) >apiMel3.Group13 5040649 96 - 8929068 ----------CAGGUAUGUGUGAGGGGGAAAAGAAAAAUAAUAAUCAG-UUAAAAAUAAAACUAAUUUUUUACUUUCUUUCUUUCUUAUAGAUAUCUUGGACAGUUU------------- ----------.((((((.((((((..(((((.((((...((.(((.((-((........)))).))).))...)))))))))..)))))).))))))..........------------- ( -23.90, z-score = -3.65, R) >triCas2.ChLG5 7512766 110 - 18847211 ----------CAAAUCUCGUUUAGAGCUUGGAGGAAGAUGAUGUUCGGGUUCUGGUUAACUGUCAUAGUCUUUUCCAUGGCAAUACUAGUCAUGGUCUACACUUACCAAUUCAAGAACUU ----------.....(((.....)))(((((((((((((.(((..(((...........))).))).)))))))(((((((.......)))))))..............))))))..... ( -23.80, z-score = 0.04, R) >consensus __________CAAUCAUCGUCCAAGGCGUGGGUUAAGAUCAUGUACGGCUUCUGGCUGUUCCUGAUCUUCUAUGCCAUGUCCAUACUGAUAUUGAUCUACACAUAUCAAUUCGACAAGUU ...............((((..((.(((.((((...((((.....((((.......)))).....)))))))).))).)).............((((........))))...))))..... ( -9.08 = -8.39 + -0.68)

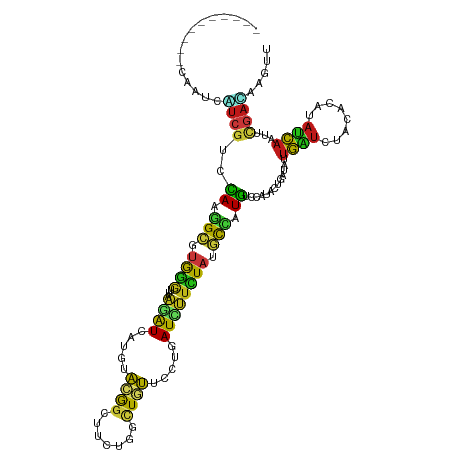
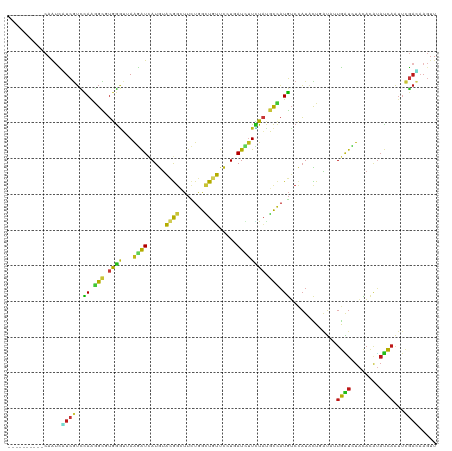
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:27 2011