
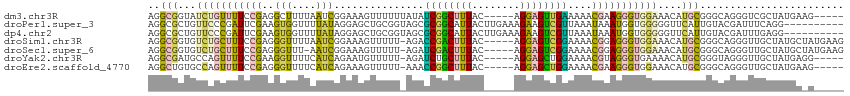
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,411,223 – 17,411,357 |
| Length | 134 |
| Max. P | 0.814655 |

| Location | 17,411,223 – 17,411,329 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 64.09 |
| Shannon entropy | 0.64510 |
| G+C content | 0.47636 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -12.65 |
| Energy contribution | -11.54 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17411223 106 + 27905053 AGGCGGUAUCUGUUUUCCGAGGCUUUUAAUCGGAAAGUUUUUUAUAUCGGCUUUAC-----AGGAGUUGAAAAACGAAGGGUGGAAACAUGCGGGCAGGGUCGCUAUGAAG----- .((((...((((((((((((.........)))))))(((((.....(((((((...-----..))))))))))))...(.(((....))).)..)))))..))))......----- ( -30.40, z-score = -1.92, R) >droPer1.super_3 6569972 106 + 7375914 AGGCGCUGUUCCCGAUUCGAAGUGGUUUUAUAGGAGCUGCGGUAGCGCGGCAUUACUUGAAAGAAGUCGUUAAAUAAAUGGUGGGGGUUCAUUGUACGAUUUCAGG---------- ...........((..(((.(((((((.........((((((....)))))))))))))))).((((((((......(((((.......)))))..)))))))).))---------- ( -25.50, z-score = 0.44, R) >dp4.chr2 23794714 106 + 30794189 AGGCGCUGUUCCCGAUUCGAAGUGGUUUUAUAGGAGCUGCGGUAGCGCGGCAUUACUUGAAAGAAGUCGUUAAAUAAAUGGUGGGGGUUCAUUGUACGAUUUGAGG---------- ..(((.((..(((..(((.(((((((.........((((((....)))))))))))))))).....(((((.....)))))...)))..)).)))...........---------- ( -23.00, z-score = 1.23, R) >droSim1.chr3R 23460481 110 - 27517382 AGGCGGUGUCUGCUUUCCGAGGGUUUUAAUCGGAAAGUUUUU-AGACCGACUUUAC-----AGGAGUCGGAAAACGGAGGGUGGAAACAUGCGGGCAGGGUUGCUAUGCUAUGAAG .(((((((((((((((((((.........)))))))(((((.-...(((((((...-----..)))))))))))).....(((....))))))))))...)))))........... ( -35.60, z-score = -2.17, R) >droSec1.super_6 280478 109 - 4358794 AGGCGGUGUCUGCUUUCCGAGGGUUU-AAUCGGAAAGUUUUU-AGAUCGACUUUAC-----AGGAGUCGGAAAACGGAGGGUGGAAACAUGCGGGCAGGGUUGCUAUGCUAUGAAG .(((((((((((((((((((......-..)))))))((((((-....((((((...-----..)))))))))))).....(((....))))))))))...)))))........... ( -34.40, z-score = -2.27, R) >droYak2.chr3R 6248610 105 + 28832112 AGGCGAUGCCAGUUUUCCGAAGGUUUUCAUCAGAAUGUUUUU-AGAUCUGCUUUAC-----AGGAGCUGGAAAACGUAGGGUGAAAACAUGCGGGUAGGGUUGCUAUGAGG----- .((((((.((.....((((...(((((((((...((((((((-......((((...-----..))))..))))))))..)))))))))...))))..))))))))......----- ( -32.80, z-score = -2.44, R) >droEre2.scaffold_4770 13494391 105 - 17746568 AGGCUGUGCCAGUUUUCCGAGGGUUUUCAUCAGAAAGUUUUU-AAACCGGCUUUAC-----AGGAGCUGGAAAACGAAGGGUGGAAACAUGCGGGCAGGGUUGCUAUGAAG----- .((((.((((.((((((((((..(((((....)))))..)))-.....(((((...-----..))))))))))))...(.(((....))).).)))).)))).........----- ( -36.30, z-score = -2.67, R) >consensus AGGCGGUGUCUGCUUUCCGAGGGUUUUAAUCAGAAAGUUUUU_AGAUCGGCUUUAC_____AGGAGUCGGAAAACGAAGGGUGGAAACAUGCGGGCAGGGUUGCUAUGA_G_____ ..(((...(((((((((((..(((....)))...............((((((((........))))))))....)))))))))))....)))........................ (-12.65 = -11.54 + -1.11)

| Location | 17,411,262 – 17,411,357 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.84 |
| Shannon entropy | 0.21488 |
| G+C content | 0.40726 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -17.92 |
| Energy contribution | -16.92 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.694790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17411262 95 + 27905053 UUUUAUAUCGGCUUUACAGGAGUUGAAAAACGAAGGGUGGAAACAUGCGGGCAGGGUCGCUAU-----GAAGCUAUAAAA-----CACUUCUAAUUAUGGAAAUG--- (((((((.(((((((.(....(((....)))...(.(((....))).)..).))))))).)))-----))))........-----((.(((((....))))).))--- ( -21.20, z-score = -1.91, R) >droSim1.chr3R 23460519 100 - 27517382 UUUUAGACCGACUUUACAGGAGUCGGAAAACGGAGGGUGGAAACAUGCGGGCAGGGUUGCUAUGCUAUGAAGCUAUAAAA-----CACUUCUAAUUAUGGAAAUG--- .......(((((((.....))))))).....((((((((....)))((...((.(((......))).))..)).......-----..))))).............--- ( -24.00, z-score = -1.88, R) >droSec1.super_6 280515 100 - 4358794 UUUUAGAUCGACUUUACAGGAGUCGGAAAACGGAGGGUGGAAACAUGCGGGCAGGGUUGCUAUGCUAUGAAGCUAUAAAA-----CACUUCUAAUUAUGGAAAUG--- .......(((((((.....))))))).....((((((((....)))((...((.(((......))).))..)).......-----..))))).............--- ( -21.20, z-score = -1.17, R) >droYak2.chr3R 6248648 98 + 28832112 UUUUAGAUCUGCUUUACAGGAGCUGGAAAACGUAGGGUGAAAACAUGCGGGUAGGGUUGCUAU-----GAGGCUAUAAAA-----CACUUCUAAUUAUGGAAAUGAGU (((((.....((((((.((.((((..(...((((..((....)).))))..)..)))).)).)-----)))))..)))))-----((.(((((....))))).))... ( -21.80, z-score = -1.23, R) >droEre2.scaffold_4770 13494429 103 - 17746568 UUUUAAACCGGCUUUACAGGAGCUGGAAAACGAAGGGUGGAAACAUGCGGGCAGGGUUGCUAU-----GAAGAUAUAAAAUAAAACACUUCUAAUUAUGGAAAUGAGU .......(((((((.....)))))))........(.(((....))).).(((......)))..-----.................((.(((((....))))).))... ( -22.00, z-score = -1.96, R) >consensus UUUUAGAUCGGCUUUACAGGAGUUGGAAAACGAAGGGUGGAAACAUGCGGGCAGGGUUGCUAU_____GAAGCUAUAAAA_____CACUUCUAAUUAUGGAAAUG___ .......(((((((.....)))))))........(.(((....))).).(((......)))........................((.(((((....))))).))... (-17.92 = -16.92 + -1.00)

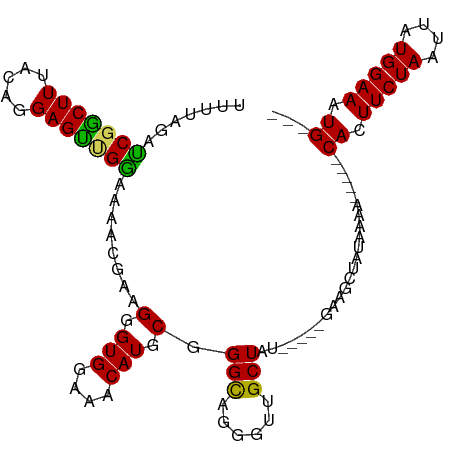
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:46 2011