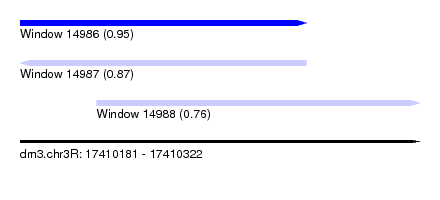
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,410,181 – 17,410,322 |
| Length | 141 |
| Max. P | 0.945996 |

| Location | 17,410,181 – 17,410,282 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Shannon entropy | 0.14386 |
| G+C content | 0.49282 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -23.56 |
| Energy contribution | -23.72 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.945996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17410181 101 + 27905053 CCUAAUCACCAGAUCUGCCAACUUAUGCCCAGAGUUGGGAUCCACCAAUUCACUG---GGCAACGAUCGUUUUAGCACCAGCAAAAAAGUGACUGAAGCUUUCG .........................(((((((((((((......)))))))..))---)))).(((..((((((((((..........))).)))))))..))) ( -30.30, z-score = -3.35, R) >droSim1.chr3R 23459404 99 - 27517382 CCUAAUCUCCAGAUCUGCCAACUCAUGCCCAGAGUUGGGAUCCACCAAUUCGCUG---GCCAACGAUCGUUUUAGCACCAGCAAA--GGUGACUGAAGCUUUCG ...........((((..(((((((.......)))))))))))..(((......))---)....(((..(((((((((((......--)))).)))))))..))) ( -30.10, z-score = -2.49, R) >droSec1.super_6 279397 99 - 4358794 CCUAAUCUCCAGAUCUGCCAACUCAUGCCCAGAGUUGGGAUCCACCAAUUCGCUG---GCCAACGAUCGUUUUAGCACCAGCAAA--GGUGACUGAAGCUUUCG ...........((((..(((((((.......)))))))))))..(((......))---)....(((..(((((((((((......--)))).)))))))..))) ( -30.10, z-score = -2.49, R) >droYak2.chr3R 6247556 99 + 28832112 CCUAAUCUCCAGAUCUGCCAACUCAUGCUCAGAGUUGGGAUCCACCAAUUUGCUGCUGUCCAACGAUCGUUUUAGCACCAGCAAA--GGUGACUGAAGCUU--- ......((.(((.....(((((((.......)))))))....((((..(((((((.(((..(((....)))...))).)))))))--)))).))).))...--- ( -29.30, z-score = -2.03, R) >droEre2.scaffold_4770 13493287 95 - 17746568 UCUAAUCUUCAGAUCUGCCAACUCAUGCCCAGAGUUGGGAUCCACCAAUUUGCUG---UCCAACGAUCGUUUU-GCACCAGCAAA--GGUGACUGAAGCUU--- ......((((((.....(((((((.......)))))))....((((..(((((((---..(((........))-)...)))))))--)))).))))))...--- ( -29.70, z-score = -2.75, R) >consensus CCUAAUCUCCAGAUCUGCCAACUCAUGCCCAGAGUUGGGAUCCACCAAUUCGCUG___GCCAACGAUCGUUUUAGCACCAGCAAA__GGUGACUGAAGCUUUCG ...........((((.((........))...(((((((......))))))).............))))(((((((((((........)))).)))))))..... (-23.56 = -23.72 + 0.16)



| Location | 17,410,181 – 17,410,282 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.46 |
| Shannon entropy | 0.14386 |
| G+C content | 0.49282 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -29.66 |
| Energy contribution | -29.34 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17410181 101 - 27905053 CGAAAGCUUCAGUCACUUUUUUGCUGGUGCUAAAACGAUCGUUGCC---CAGUGAAUUGGUGGAUCCCAACUCUGGGCAUAAGUUGGCAGAUCUGGUGAUUAGG ..........(((((((..((((((..(.....(((....)))(((---(((.(..((((......))))).))))))....)..))))))...)))))))... ( -30.40, z-score = -1.14, R) >droSim1.chr3R 23459404 99 + 27517382 CGAAAGCUUCAGUCACC--UUUGCUGGUGCUAAAACGAUCGUUGGC---CAGCGAAUUGGUGGAUCCCAACUCUGGGCAUGAGUUGGCAGAUCUGGAGAUUAGG ......(((((..((((--(((((((((.....(((....))).))---)))))))..))))(((((((((((.......)))))))..)))))))))...... ( -35.40, z-score = -1.98, R) >droSec1.super_6 279397 99 + 4358794 CGAAAGCUUCAGUCACC--UUUGCUGGUGCUAAAACGAUCGUUGGC---CAGCGAAUUGGUGGAUCCCAACUCUGGGCAUGAGUUGGCAGAUCUGGAGAUUAGG ......(((((..((((--(((((((((.....(((....))).))---)))))))..))))(((((((((((.......)))))))..)))))))))...... ( -35.40, z-score = -1.98, R) >droYak2.chr3R 6247556 99 - 28832112 ---AAGCUUCAGUCACC--UUUGCUGGUGCUAAAACGAUCGUUGGACAGCAGCAAAUUGGUGGAUCCCAACUCUGAGCAUGAGUUGGCAGAUCUGGAGAUUAGG ---...(((((..((((--(((((((.((((((........)))).)).)))))))..))))(((((((((((.......)))))))..)))))))))...... ( -36.10, z-score = -2.53, R) >droEre2.scaffold_4770 13493287 95 + 17746568 ---AAGCUUCAGUCACC--UUUGCUGGUGC-AAAACGAUCGUUGGA---CAGCAAAUUGGUGGAUCCCAACUCUGGGCAUGAGUUGGCAGAUCUGAAGAUUAGA ---...(((((..((((--(((((((.(.(-((........))).)---)))))))..))))(((((((((((.......)))))))..)))))))))...... ( -32.30, z-score = -1.92, R) >consensus CGAAAGCUUCAGUCACC__UUUGCUGGUGCUAAAACGAUCGUUGGC___CAGCGAAUUGGUGGAUCCCAACUCUGGGCAUGAGUUGGCAGAUCUGGAGAUUAGG ......(((((..((((..(((((((.......(((....)))......)))))))..))))(((((((((((.......)))))))..)))))))))...... (-29.66 = -29.34 + -0.32)


| Location | 17,410,208 – 17,410,322 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.69 |
| Shannon entropy | 0.22682 |
| G+C content | 0.45870 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -20.02 |
| Energy contribution | -21.18 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.764195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17410208 114 + 27905053 CCCAGAGUUGGGAUCCACCAAUUCACU---GGGCAACGAUCGUUUUAGCACCAGCAAAAAAGUGACUGAAGCUUUCGAGCUCGAGCACCUGAAUAGUUUUUUUUUUUUUUUUGUCCC ....(((((((......)))))))...---(((((((((..((((((((((..........))).)))))))..))).((....))........................)))))). ( -29.90, z-score = -1.72, R) >droSim1.chr3R 23459431 100 - 27517382 CCCAGAGUUGGGAUCCACCAAUUCGCU---GGCCAACGAUCGUUUUAGCACCAGCAAA--GGUGACUGAAGCUUUCGAGCUCGAGCACCUAAAUAGUUUUAUUUC------------ ....(((((((......)))))))(((---(((...(((..(((((((((((......--)))).)))))))..))).)))..)))...................------------ ( -31.60, z-score = -3.12, R) >droSec1.super_6 279424 100 - 4358794 CCCAGAGUUGGGAUCCACCAAUUCGCU---GGCCAACGAUCGUUUUAGCACCAGCAAA--GGUGACUGAAGCUUUCGAGCUCGAGCACCUAAAUAGUUUUAUUUC------------ ....(((((((......)))))))(((---(((...(((..(((((((((((......--)))).)))))))..))).)))..)))...................------------ ( -31.60, z-score = -3.12, R) >droYak2.chr3R 6247583 97 + 28832112 CUCAGAGUUGGGAUCCACCAAUUUGCUGCUGUCCAACGAUCGUUUUAGCACCAGCAAA--GGUGACUGAAGCUUU------CGAGCACCUAAAUACUUUUAUUUC------------ ....(((((((......)))))))..((((......(((..(((((((((((......--)))).)))))))..)------))))))..................------------ ( -25.40, z-score = -1.86, R) >droEre2.scaffold_4770 13493314 93 - 17746568 CCCAGAGUUGGGAUCCACCAAUUUGCU---GUCCAACGAUCGUUUU-GCACCAGCAAA--GGUGACUGAAGCUUU------CGAGCACCUAAAUAGUUUUAUUUC------------ ....(((((((......)))))))(((---((............((-((....))))(--((((..(((.....)------))..)))))..)))))........------------ ( -20.10, z-score = -0.46, R) >consensus CCCAGAGUUGGGAUCCACCAAUUCGCU___GGCCAACGAUCGUUUUAGCACCAGCAAA__GGUGACUGAAGCUUUCGAGCUCGAGCACCUAAAUAGUUUUAUUUC____________ ....(((((((......)))))))(((.........(((..(((((((((((........)))).)))))))..)))......)))............................... (-20.02 = -21.18 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:44 2011