
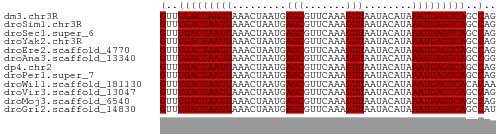
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,385,382 – 17,385,438 |
| Length | 56 |
| Max. P | 0.982853 |

| Location | 17,385,382 – 17,385,438 |
|---|---|
| Length | 56 |
| Sequences | 12 |
| Columns | 56 |
| Reading direction | forward |
| Mean pairwise identity | 98.54 |
| Shannon entropy | 0.03675 |
| G+C content | 0.30060 |
| Mean single sequence MFE | -9.81 |
| Consensus MFE | -9.60 |
| Energy contribution | -9.61 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17385382 56 + 27905053 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.30, R) >droSim1.chr3R 23433213 56 - 27517382 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.30, R) >droSec1.super_6 254623 56 - 4358794 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.30, R) >droYak2.chr3R 6222574 56 + 28832112 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.30, R) >droEre2.scaffold_4770 13466454 56 - 17746568 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.30, R) >droAna3.scaffold_13340 17722142 56 + 23697760 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCGG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -1.88, R) >dp4.chr2 3686235 56 + 30794189 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.30, R) >droPer1.super_7 3781216 56 - 4445127 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.30, R) >droWil1.scaffold_181130 4179915 56 - 16660200 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCACACAA ((.(((((((((.........(((.......)))........))))))))).)).. ( -10.73, z-score = -2.90, R) >droVir3.scaffold_13047 4690367 56 - 19223366 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.30, R) >droMoj3.scaffold_6540 16172837 56 - 34148556 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.30, R) >droGri2.scaffold_14830 5436621 56 + 6267026 GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAU (.((((((((((.........(((.......)))........)))))))))).).. ( -9.73, z-score = -2.43, R) >consensus GUUUGACUAAUUAAACUAAUGAGCGUUCAAAGUUAAUACAUAAAUUAGUCAGCCAG (..(((((((((.........(((.......)))........)))))))))..).. ( -9.60 = -9.61 + 0.00)
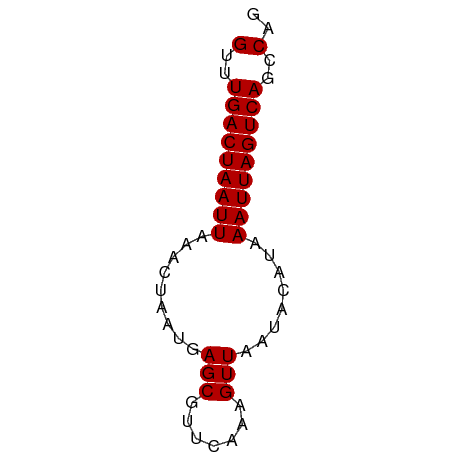
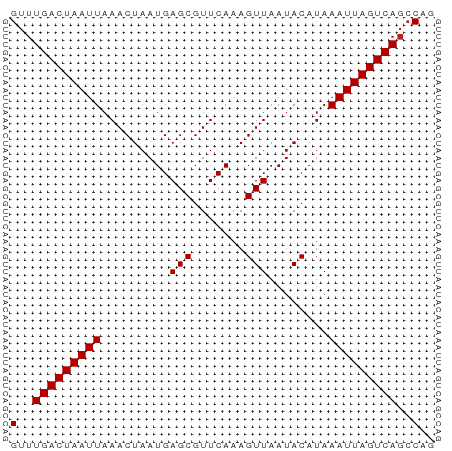
| Location | 17,385,382 – 17,385,438 |
|---|---|
| Length | 56 |
| Sequences | 12 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 98.54 |
| Shannon entropy | 0.03675 |
| G+C content | 0.30060 |
| Mean single sequence MFE | -9.91 |
| Consensus MFE | -9.91 |
| Energy contribution | -9.91 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17385382 56 - 27905053 CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.86, R) >droSim1.chr3R 23433213 56 + 27517382 CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.86, R) >droSec1.super_6 254623 56 + 4358794 CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.86, R) >droYak2.chr3R 6222574 56 - 28832112 CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.86, R) >droEre2.scaffold_4770 13466454 56 + 17746568 CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.86, R) >droAna3.scaffold_13340 17722142 56 - 23697760 CCGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.92, R) >dp4.chr2 3686235 56 - 30794189 CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.86, R) >droPer1.super_7 3781216 56 + 4445127 CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.86, R) >droWil1.scaffold_181130 4179915 56 + 16660200 UUGUGUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -10.00, z-score = -1.69, R) >droVir3.scaffold_13047 4690367 56 + 19223366 CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.86, R) >droMoj3.scaffold_6540 16172837 56 + 34148556 CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.86, R) >droGri2.scaffold_14830 5436621 56 - 6267026 AUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.90, z-score = -1.90, R) >consensus CUGGCUGACUAAUUUAUGUAUUAACUUUGAACGCUCAUUAGUUUAAUUAGUCAAAC .....(((((((((........((((.(((....)))..)))).)))))))))... ( -9.91 = -9.91 + -0.00)

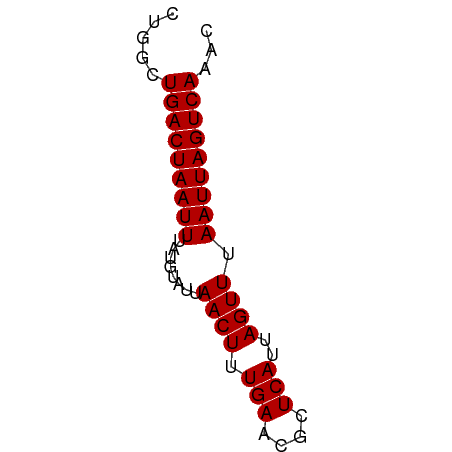
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:40 2011