
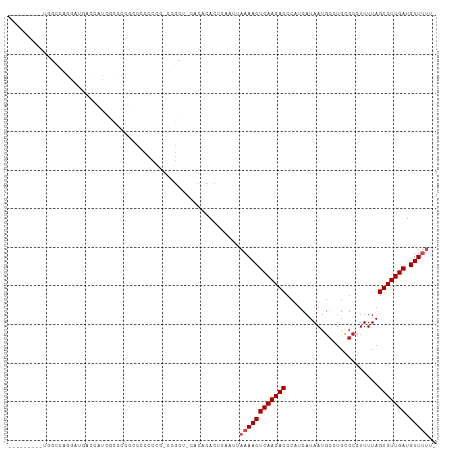
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,364,718 – 17,364,849 |
| Length | 131 |
| Max. P | 0.797882 |

| Location | 17,364,718 – 17,364,817 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.31 |
| Shannon entropy | 0.59103 |
| G+C content | 0.46189 |
| Mean single sequence MFE | -15.98 |
| Consensus MFE | -7.89 |
| Energy contribution | -8.25 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.593230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17364718 99 + 27905053 ---------UGGCCAGGAUGACCCCCGCCUCGUCCUCUCA-CCGCU-CACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUUU- ---------.((..(((((((........)))))))....-))...-.............((((((((((((.......((.((....)).))...))))))).))))).- ( -18.00, z-score = -1.37, R) >droSim1.chrU 7260733 99 + 15797150 ---------UGGCCAGGAUGACCCCCGUCUCGCCCUCCCG-CCGCU-CACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUUU- ---------.(((.(((..(((....)))....)))...)-))...-.............((((((((((((.......((.((....)).))...))))))).))))).- ( -17.10, z-score = -0.74, R) >droSec1.super_6 233539 99 - 4358794 ---------UGGCCAGGAUGACCACCGCCUCGCCCUCCCG-CCGCU-CACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUUU- ---------.(((.(((.((.....))))).)))......-.....-.............((((((((((((.......((.((....)).))...))))))).))))).- ( -16.90, z-score = -0.53, R) >droYak2.chr3R 6194154 93 + 28832112 ---------UGGCCAGGAUGACCCUCGCCUCGCCCUUUCC-CCGCU-CACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCU------UUUAGUCUUGAUGUUUUU- ---------.(((.(((.((.....))))).)))......-.....-.............((((((((((((..............------....))))))).))))).- ( -17.37, z-score = -2.09, R) >droEre2.scaffold_4770 13444664 99 - 17746568 ---------UGGCCAGGAUAACCCCCGCGUCGCCCUUUCC-CCACU-CACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUUU- ---------.((((.((......)).).))).........-.....-.............((((((((((((.......((.((....)).))...))))))).))))).- ( -13.30, z-score = -0.13, R) >dp4.chr2 3665640 106 + 30794189 -UUGUCGGAGUCCUAGCCCCCCCAUCCCGCUGCCACUCUU-UCGGC-CACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUAC-- -..((((((((..((((...........))))..))))..-.))))-...............((((((((((.......((.((....)).))...))))))).)))..-- ( -17.00, z-score = -0.62, R) >droPer1.super_7 3760523 106 - 4445127 -UUGUCGGAGUCCUAGCACCCCCAUCCCGCUGCCACUCUU-CCGGC-CACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUAC-- -..((((((((..((((...........))))..))...)-)))))-...............((((((((((.......((.((....)).))...))))))).)))..-- ( -18.30, z-score = -0.96, R) >droWil1.scaffold_181130 4150720 79 - 16660200 ------------------------------UGCCCAAAAU-GCUCU-CACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCAUGUGCCUUUUAGUCUUGAUGUUUUUU ------------------------------..........-.....-.............((((((((((((....((((.....)))).......))))))).))))).. ( -10.20, z-score = -0.82, R) >droVir3.scaffold_13047 4669665 97 - 19223366 --------CUGCUGUGCUUUUUUAUUGGACACCCACCCAG--CGG--CACACACUCAAUUAAAACUCAAGACCCUUCAUAAUGCAUGCGCCUUUUAGUCUUGAUGUUUU-- --------.((.((((((.......(((....))).....--.))--)))))).......((((((((((((.......((.((....)).))...))))))).)))))-- ( -18.72, z-score = -1.06, R) >droGri2.scaffold_14830 5415395 96 + 6267026 ---------UUUUUCUGUUUUUUUUUUUUCGCCCACCCAG--CGA--CACACACUCAAUUAAAACUCAAGACCCUUCAUAAUGCAUGCGCCUUUUAGUCUUGAUGUUUU-- ---------...................((((.......)--)))--.............((((((((((((.......((.((....)).))...))))))).)))))-- ( -13.10, z-score = -1.86, R) >droMoj3.scaffold_6540 16151038 109 - 34148556 UCGUUUUGCUUUUUUUUUUUUUCGGUGUACGCCCACCCAGUCCGAUGCACACACUCAAUUAAAACUCAAGACCCUUCAUAAUGCAUGCGCCUUUUAGUCUUGAUGUUUU-- ........................(((((((..(.....)..)).)))))..........((((((((((((.......((.((....)).))...))))))).)))))-- ( -15.80, z-score = -0.34, R) >consensus _________UGGCCAGGAUGACCAUCGCCCCGCCCCCCCG_CCGCU_CACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUUU_ ............................................................((((((((((((........................))))))).))))).. ( -7.89 = -8.25 + 0.36)


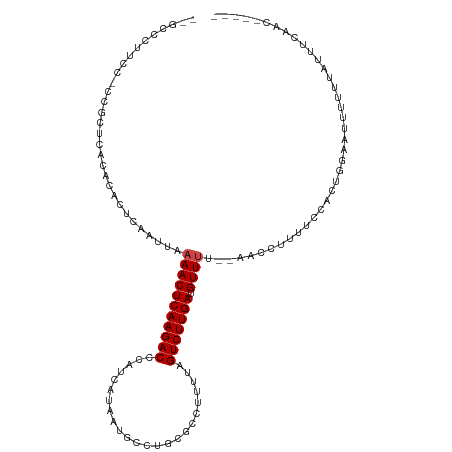
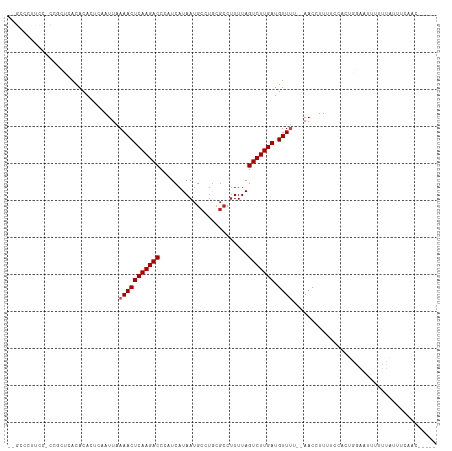
| Location | 17,364,740 – 17,364,849 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.65 |
| Shannon entropy | 0.53965 |
| G+C content | 0.39814 |
| Mean single sequence MFE | -13.48 |
| Consensus MFE | -7.48 |
| Energy contribution | -7.64 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17364740 109 + 27905053 --GUCCUCUCACCGCUCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUUU-AACCUUUUCCCCUGGAAUUUUUUAUUUCAACA---- --...........................((((((((((((.......((.((....)).))...))))))).))))).-............((((((.....))))))...---- ( -11.60, z-score = -1.14, R) >droSim1.chrU 7260755 108 + 15797150 --GCCCUCCCGCCGCUCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUUU-AACCUUUUCCCCUGGAAUUUUUUAUUUCAAC----- --...........................((((((((((((.......((.((....)).))...))))))).))))).-............((((((.....))))))..----- ( -11.60, z-score = -0.91, R) >droSec1.super_6 233561 108 - 4358794 --GCCCUCCCGCCGCUCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUUU-AACCUUUUCCCCUGGAAUUUUUUAUUUCAAC----- --...........................((((((((((((.......((.((....)).))...))))))).))))).-............((((((.....))))))..----- ( -11.60, z-score = -0.91, R) >droYak2.chr3R 6194176 102 + 28832112 --GCCCUUUCCCCGCUCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCU------UUUAGUCUUGAUGUUUUU-AACCUUUUCCGCUGGAAUUUUUUAUUUCAAC----- --.....((((..((..............((((((((((((..............------....))))))).))))).-..........)).))))..............----- ( -12.28, z-score = -1.91, R) >droEre2.scaffold_4770 13444686 108 - 17746568 --GCCCUUUCCCCACUCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUUU-AACCUUUUCCACUGGAAUUUUGUAUUUCAAC----- --...........................((((((((((((.......((.((....)).))...))))))).))))).-............((((((.....))))))..----- ( -11.60, z-score = -0.98, R) >droAna3.scaffold_13340 17702928 107 + 23697760 --ACCCCUUUUUUGGCCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCGACGCCUUUUAGUCUUGAUGUUUC--AACCUUUUCGACAAGAAUUUUUUAUUUCAAC----- --...((......))...............(((((((((((.......((.((....)).))...))))))).)))).--..............(((........)))...----- ( -9.60, z-score = -0.04, R) >dp4.chr2 3665668 109 + 30794189 CUGCCACUCUUUCGGCCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUAC--AACCUUUUCGACACUUUUUUCGUUUUUUGGU----- ..(((........)))...............((((((((((.......((.((....)).))...))))))).)))..--.(((....(((........)))......)))----- ( -14.00, z-score = -0.15, R) >droPer1.super_7 3760551 102 - 4445127 CUGCCACUCUUCCGGCCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUAC--AACCUUUUCGACACUUUUUUUGGU------------ ..(((........)))...............((((((((((.......((.((....)).))...))))))).)))..--.(((.................)))------------ ( -11.63, z-score = 0.37, R) >droWil1.scaffold_181130 4150730 105 - 16660200 -----------GCUCUCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCAUGUGCCUUUUAGUCUUGAUGUUUUUUAACCUUUUUUCCCCGAUUUAUUUGGGACGUUUUUUU -----------..................((((((((((((....((((.....)))).......))))))).))))).............(((((.....))))).......... ( -16.40, z-score = -1.72, R) >droVir3.scaffold_13047 4669696 93 - 19223366 ------------GCGGCACACACUCAAUUAAAACUCAAGACCCUUCAUAAUGCAUGCGCCUUUUAGUCUUGAUGUUUU--AACCUUUUCUUUUUUUUUCCCAGCUGC--------- ------------(((((..........((((((((((((((.......((.((....)).))...))))))).)))))--))............(......))))))--------- ( -15.20, z-score = -1.65, R) >droGri2.scaffold_14830 5415425 92 + 6267026 ------------GCGACACACACUCAAUUAAAACUCAAGACCCUUCAUAAUGCAUGCGCCUUUUAGUCUUGAUGUUUU--AACCUUUUCGUUUUUUUGUGU-GUUGC--------- ------------((((((((((......(((((((((((((.......((.((....)).))...))))))).)))))--)((......)).....)))))-)))))--------- ( -23.90, z-score = -4.25, R) >droMoj3.scaffold_6540 16151077 92 - 34148556 --------GUCCGAUGCACACACUCAAUUAAAACUCAAGACCCUUCAUAAUGCAUGCGCCUUUUAGUCUUGAUGUUUU--AACCUUUCCCUUUUUU-----AGCUGC--------- --------...................((((((((((((((.......((.((....)).))...))))))).)))))--))..............-----......--------- ( -12.40, z-score = -1.07, R) >consensus __GCCCUUCC_CCGCUCACACACUCAAUUAAAACUCAAGACCCAUCAUAAUGCCUGCGCCUUUUAGUCUUGAUGUUUU__AACCUUUUCCACUGGAAUUUUUUAUUUCAAC_____ ..............................(((((((((((........................))))))).))))....................................... ( -7.48 = -7.64 + 0.17)

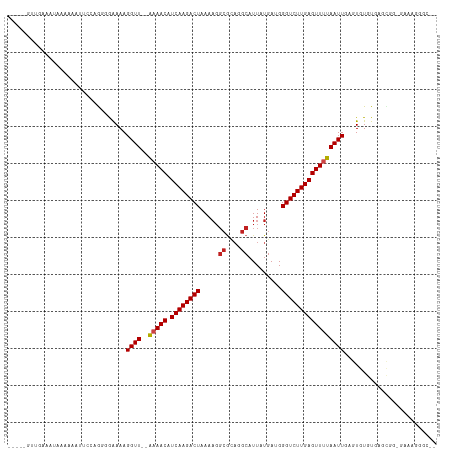
| Location | 17,364,740 – 17,364,849 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.65 |
| Shannon entropy | 0.53965 |
| G+C content | 0.39814 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17364740 109 - 27905053 ----UGUUGAAAUAAAAAAUUCCAGGGGAAAAGGUU-AAAAACAUCAAGACUAAAAGGCGCAGGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGAGCGGUGAGAGGAC-- ----.(((.(.(((.....((((...))))..((((-((((...((((((((.....((....)).........)))))))).))))))))...))).).)))...........-- ( -18.94, z-score = -0.31, R) >droSim1.chrU 7260755 108 - 15797150 -----GUUGAAAUAAAAAAUUCCAGGGGAAAAGGUU-AAAAACAUCAAGACUAAAAGGCGCAGGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGAGCGGCGGGAGGGC-- -----((((..(((.....((((...))))..((((-((((...((((((((.....((....)).........)))))))).)))))))).....)))...))))........-- ( -20.74, z-score = -0.56, R) >droSec1.super_6 233561 108 + 4358794 -----GUUGAAAUAAAAAAUUCCAGGGGAAAAGGUU-AAAAACAUCAAGACUAAAAGGCGCAGGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGAGCGGCGGGAGGGC-- -----((((..(((.....((((...))))..((((-((((...((((((((.....((....)).........)))))))).)))))))).....)))...))))........-- ( -20.74, z-score = -0.56, R) >droYak2.chr3R 6194176 102 - 28832112 -----GUUGAAAUAAAAAAUUCCAGCGGAAAAGGUU-AAAAACAUCAAGACUAAA------AGGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGAGCGGGGAAAGGGC-- -----..............((((.(((.(...((((-((((...((((((((...------.(......)....)))))))).))))))))...).))).....))))......-- ( -20.40, z-score = -1.62, R) >droEre2.scaffold_4770 13444686 108 + 17746568 -----GUUGAAAUACAAAAUUCCAGUGGAAAAGGUU-AAAAACAUCAAGACUAAAAGGCGCAGGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGAGUGGGGAAAGGGC-- -----..............(((((.(.(....((((-((((...((((((((.....((....)).........)))))))).)))))))).......).).))))).......-- ( -20.64, z-score = -0.79, R) >droAna3.scaffold_13340 17702928 107 - 23697760 -----GUUGAAAUAAAAAAUUCUUGUCGAAAAGGUU--GAAACAUCAAGACUAAAAGGCGUCGGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGGCCAAAAAAGGGGU-- -----..............(((((((((....((((--(((((.((((((((......(((((......)))))))))))))))))))))).......)))).....)))))..-- ( -22.00, z-score = -1.21, R) >dp4.chr2 3665668 109 - 30794189 -----ACCAAAAAACGAAAAAAGUGUCGAAAAGGUU--GUAACAUCAAGACUAAAAGGCGCAGGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGGCCGAAAGAGUGGCAG -----..................(((((....(((.--.((...((((((((..(((((.((..(.....).)).))))).))))....))))...))..))).......))))). ( -22.50, z-score = -0.33, R) >droPer1.super_7 3760551 102 + 4445127 ------------ACCAAAAAAAGUGUCGAAAAGGUU--GUAACAUCAAGACUAAAAGGCGCAGGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGGCCGGAAGAGUGGCAG ------------...........(((((....(((.--.((...((((((((..(((((.((..(.....).)).))))).))))....))))...))..))).......))))). ( -22.50, z-score = -0.48, R) >droWil1.scaffold_181130 4150730 105 + 16660200 AAAAAAACGUCCCAAAUAAAUCGGGGAAAAAAGGUUAAAAAACAUCAAGACUAAAAGGCACAUGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGAGAGC----------- .........((((..........)))).....((((((((....((((((((.....((....)).........)))))))).))))))))......((....))----------- ( -19.84, z-score = -1.14, R) >droVir3.scaffold_13047 4669696 93 + 19223366 ---------GCAGCUGGGAAAAAAAAAGAAAAGGUU--AAAACAUCAAGACUAAAAGGCGCAUGCAUUAUGAAGGGUCUUGAGUUUUAAUUGAGUGUGUGCCGC------------ ---------...((.((.(.......(.(...((((--(((((.((((((((.....((....)).........)))))))))))))))))...).).).))))------------ ( -18.24, z-score = -0.18, R) >droGri2.scaffold_14830 5415425 92 - 6267026 ---------GCAAC-ACACAAAAAAACGAAAAGGUU--AAAACAUCAAGACUAAAAGGCGCAUGCAUUAUGAAGGGUCUUGAGUUUUAAUUGAGUGUGUGUCGC------------ ---------((.((-(((((............((((--(((((.((((((((.....((....)).........)))))))))))))))))...))))))).))------------ ( -27.10, z-score = -3.92, R) >droMoj3.scaffold_6540 16151077 92 + 34148556 ---------GCAGCU-----AAAAAAGGGAAAGGUU--AAAACAUCAAGACUAAAAGGCGCAUGCAUUAUGAAGGGUCUUGAGUUUUAAUUGAGUGUGUGCAUCGGAC-------- ---------((((((-----............((((--(((((.((((((((.....((....)).........))))))))))))))))).)))...))).......-------- ( -19.66, z-score = -1.15, R) >consensus _____GUUGAAAUAAAAAAUUCCAGGGGAAAAGGUU__AAAACAUCAAGACUAAAAGGCGCAGGCAUUAUGAUGGGUCUUGAGUUUUAAUUGAGUGUGUGAGCGG_GAAAGGGC__ ......................................(((((.((((((((.....((....)).........)))))))))))))............................. (-12.69 = -12.83 + 0.15)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:32 2011