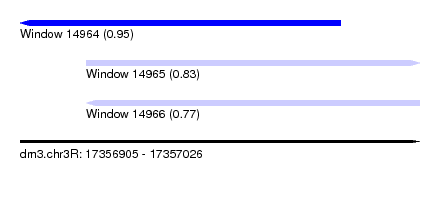
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,356,905 – 17,357,026 |
| Length | 121 |
| Max. P | 0.950399 |

| Location | 17,356,905 – 17,357,002 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.03 |
| Shannon entropy | 0.35117 |
| G+C content | 0.39929 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -18.04 |
| Energy contribution | -17.92 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17356905 97 - 27905053 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGC--------------UAUAGCUAUGG--CCACAGGCCACGC- ....(((((........)))))((((.(((((((((.......))))))))).))))((.((((.((((((((--------------....))....)--))))).)))).))- ( -28.60, z-score = -2.40, R) >droSim1.chr3R 23404997 97 + 27517382 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGC--------------UAUAGCUAUGG--CCACAGGCCACGC- ....(((((........)))))((((.(((((((((.......))))))))).))))((.((((.((((((((--------------....))....)--))))).)))).))- ( -28.60, z-score = -2.40, R) >droSec1.super_6 225724 97 + 4358794 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCAGUUUCCGAUUGUUUUGCGUGUGGUUC--------------UAUAGCUAUGG--CCACAGGCCACGC- .(((((...(((...(((((((.....(((((((((.......))))))))))))))))...)))..))))).--------------........(((--((...)))))...- ( -25.00, z-score = -2.15, R) >droYak2.chr3R 6186730 111 - 28832112 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGCUAUAUAUCUAUAGCUAUAGCUAUGG--CCACAGGCCACGC- ....(((((........)))))((((.(((((((((.......))))))))).))))((.((((.(((((..........(((((((....)))))))--))))).)))).))- ( -32.90, z-score = -2.97, R) >droEre2.scaffold_4770 13437075 109 + 17746568 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGCUAUAGAUACUCG--UAUAGCUAUGG--CCACAGGCCAUGC- ....(((((........)))))((((.(((((((((.......))))))))).))))((.((((.(((((((((((((.....).--))))))....)--))))).)))).))- ( -33.10, z-score = -3.10, R) >droAna3.scaffold_13340 17695440 91 - 23697760 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGU--------------UCAUCGGCCGCAGGCCACUC--------- ....(((((........)))))((((.(((((((((.......))))))))).))))...((((.((((((--------------......)))))).))))...--------- ( -27.40, z-score = -2.34, R) >dp4.chr2 3657475 102 - 30794189 GAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGC-----------AGCAGCAGCCACAGGCCAAUGGGCCACGC- (..(((.........(((((((.....(((((((((.......))))))))))))))))(((((.((((((((-----------....))).))))).))))))))..)....- ( -33.30, z-score = -2.28, R) >droPer1.super_7 3752602 102 + 4445127 GAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGC-----------AGCAGCAGCCACAGGCCAAUGGGCCACGC- (..(((.........(((((((.....(((((((((.......))))))))))))))))(((((.((((((((-----------....))).))))).))))))))..)....- ( -33.30, z-score = -2.28, R) >droWil1.scaffold_181130 4138747 100 + 16660200 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCAAUUGUUUGGCAUGCGGU--------------GCAAUAGCCACAGGACGAAACGCUAUCUC ...((....((((..(((((((.....(((((((((.......))))))))))))))))..))))((.(((--------------......))).))))............... ( -20.20, z-score = -0.26, R) >droVir3.scaffold_13047 4659768 92 + 19223366 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCAAUUGUUUGGCGUGUGGU--------------GCAACAGCCACAGGCCAACGG-------- ...((..........(((((((.....(((((((((.......))))))))))))))))(((((.((((((--------------......)))))).))))).))-------- ( -28.90, z-score = -2.84, R) >droMoj3.scaffold_6540 16138993 92 + 34148556 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGU--------------GCAAUAGCCGCAGGCCAAAGG-------- ....(((((........)))))((((.(((((((((.......))))))))).)))).((((((.((((((--------------......)))))).))))))..-------- ( -29.70, z-score = -3.30, R) >droGri2.scaffold_14830 5406038 92 - 6267026 AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGU--------------GCAAUAGCCACCGGCCAAUGG-------- ...(((.........(((((((.....(((((((((.......))))))))))))))))(((((..(((((--------------......)))))..))))))))-------- ( -24.60, z-score = -1.49, R) >consensus AAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGC____________GCAAUAGCCACAGGCCAACAGGCCACGC_ ..((((...((((..(((((((.....(((((((((.......))))))))))))))))..))))..))))........................................... (-18.04 = -17.92 + -0.12)

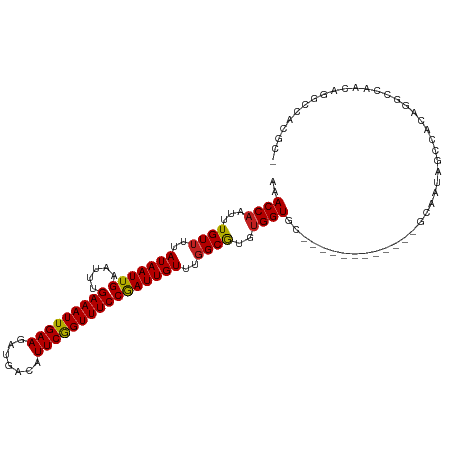
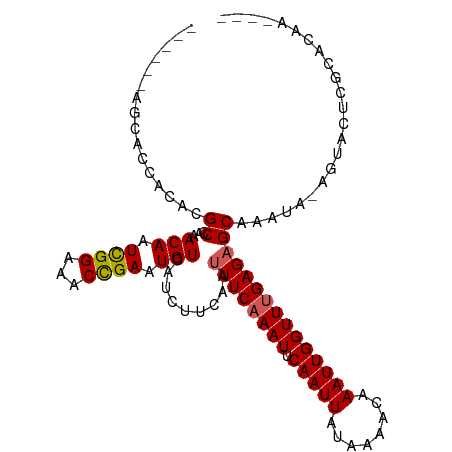
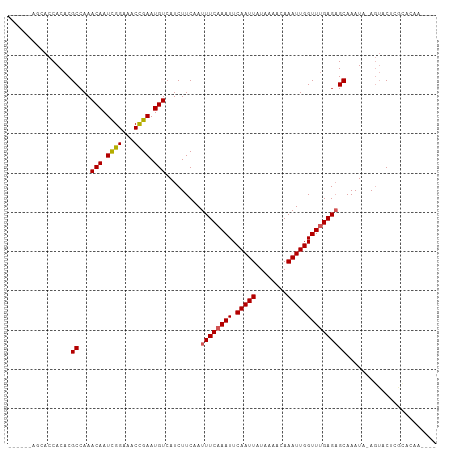
| Location | 17,356,925 – 17,357,026 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Shannon entropy | 0.24391 |
| G+C content | 0.35014 |
| Mean single sequence MFE | -14.58 |
| Consensus MFE | -12.88 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17356925 101 + 27905053 ---UAUAGCACCACACGCCAAACAAUCGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUUGAGAGCAAAUA-AGUACUCGCACAA---- ---....((............(((.((((...)))).))).........((((((((.(((((........))))))))))))).......-.......))....---- ( -15.40, z-score = -1.72, R) >droSim1.chr3R 23405017 101 - 27517382 ---UAUAGCACCACACGCCAAACAAUCGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUUGAGAGCAAAUA-AGUACUCGCACAA---- ---....((............(((.((((...)))).))).........((((((((.(((((........))))))))))))).......-.......))....---- ( -15.40, z-score = -1.72, R) >droSec1.super_6 225744 101 - 4358794 ---UAUAGAACCACACGCAAAACAAUCGGAAACUGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUUGAGAGCAAAUA-AGUACUCGCACAA---- ---.............((...(((.(((....).)).))).........((((((((.(((((........))))))))))))).......-.......))....---- ( -13.90, z-score = -1.03, R) >droYak2.chr3R 6186761 104 + 28832112 AUAUAUAGCACCACACGCCAAACAAUCGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUUGAGAGCAAAUA-AGUACUCACACAA---- .......((............(((.((((...)))).))).........((((((((.(((((........))))))))))))))).....-.............---- ( -15.30, z-score = -2.03, R) >droEre2.scaffold_4770 13437104 104 - 17746568 AUCUAUAGCACCACACGCCAAACAAUCGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUUGAGAGCAAAUA-AGUACUCGCACAA---- .......((............(((.((((...)))).))).........((((((((.(((((........))))))))))))).......-.......))....---- ( -15.40, z-score = -1.57, R) >droAna3.scaffold_13340 17695460 99 + 23697760 ---------ACCACACGCCAAACAAUCGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUUGAGAGCAAAUA-AGUGCGAGCGCACACAC ---------.......((...(((.((((...)))).))).........((((((((.(((((........))))))))))))))).....-.((((....)))).... ( -20.50, z-score = -2.54, R) >dp4.chr2 3657503 92 + 30794189 ------UGCACCACACGCCAAACAAUCGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUCGAGAGCAAAUACAGAGCG----------- ------(((............(((.((((...)))).))).........((((.(((.(((((........)))))))).)))))))...........----------- ( -11.70, z-score = -0.26, R) >droPer1.super_7 3752630 92 - 4445127 ------UGCACCACACGCCAAACAAUCGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUCGAGAGCAAAUACAGAGCG----------- ------(((............(((.((((...)))).))).........((((.(((.(((((........)))))))).)))))))...........----------- ( -11.70, z-score = -0.26, R) >droVir3.scaffold_13047 4659789 95 - 19223366 ---------ACCACACGCCAAACAAUUGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUUGAGCGCAAAUA-AAUACAGAUACAC---- ---------......((((((((..((((((...(((.......)))..))))))...(((((........)))))))))).)))......-.............---- ( -12.70, z-score = -1.15, R) >droGri2.scaffold_14830 5406059 95 + 6267026 ---------ACCACACGCCAAACAAUCGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUUGAGCGCAAAUA-AAUACACACACAC---- ---------......(((...(((.((((...)))).)))...........((((((.(((((........))))))))))))))......-.............---- ( -13.80, z-score = -2.19, R) >consensus ______AGCACCACACGCCAAACAAUCGGAAACCGAAUGUCAUCUUCAAUUUCAAAUUCAAUUAUAAAACAAAUUGGUUUGAGAGCAAAUA_AGUACUCGCACAA____ ................((...(((.((((...)))).))).........((((((((.(((((........)))))))))))))))....................... (-12.88 = -13.10 + 0.22)
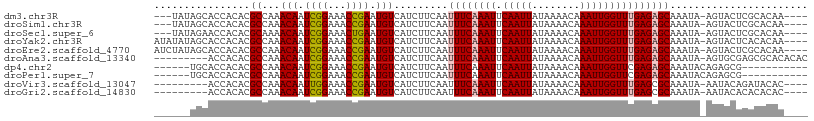
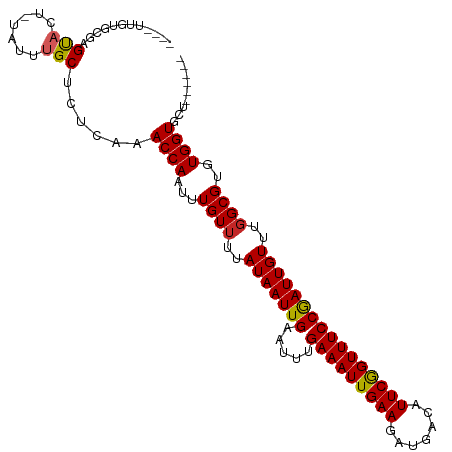
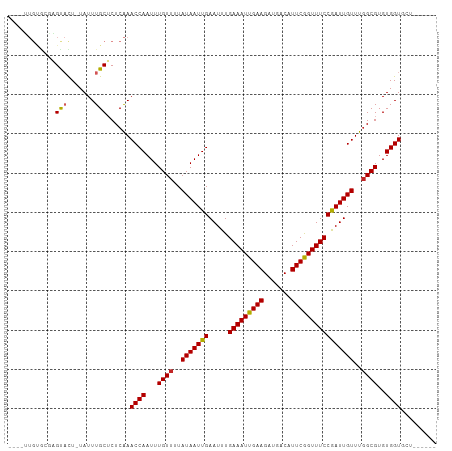
| Location | 17,356,925 – 17,357,026 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Shannon entropy | 0.24391 |
| G+C content | 0.35014 |
| Mean single sequence MFE | -23.24 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.16 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17356925 101 - 27905053 ----UUGUGCGAGUACU-UAUUUGCUCUCAAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGCUAUA--- ----....(((((((..-....)))))....((((...((((..(((((((.....(((((((((.......))))))))))))))))..))))..))))))....--- ( -24.60, z-score = -2.11, R) >droSim1.chr3R 23405017 101 + 27517382 ----UUGUGCGAGUACU-UAUUUGCUCUCAAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGCUAUA--- ----....(((((((..-....)))))....((((...((((..(((((((.....(((((((((.......))))))))))))))))..))))..))))))....--- ( -24.60, z-score = -2.11, R) >droSec1.super_6 225744 101 + 4358794 ----UUGUGCGAGUACU-UAUUUGCUCUCAAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCAGUUUCCGAUUGUUUUGCGUGUGGUUCUAUA--- ----...((.(((((..-....))))).))(((((...(((...(((((((.....(((((((((.......))))))))))))))))...)))..))))).....--- ( -23.70, z-score = -2.50, R) >droYak2.chr3R 6186761 104 - 28832112 ----UUGUGUGAGUACU-UAUUUGCUCUCAAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGCUAUAUAU ----...((.(((((..-....))))).)).((((...((((..(((((((.....(((((((((.......))))))))))))))))..))))..))))......... ( -24.30, z-score = -2.13, R) >droEre2.scaffold_4770 13437104 104 + 17746568 ----UUGUGCGAGUACU-UAUUUGCUCUCAAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGCUAUAGAU ----(((((((((((..-....)))))....((((...((((..(((((((.....(((((((((.......))))))))))))))))..))))..)))))).)))).. ( -25.00, z-score = -1.87, R) >droAna3.scaffold_13340 17695460 99 - 23697760 GUGUGUGCGCUCGCACU-UAUUUGCUCUCAAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGU--------- (((((......))))).-.............((((...((((..(((((((.....(((((((((.......))))))))))))))))..))))..))))--------- ( -23.40, z-score = -0.98, R) >dp4.chr2 3657503 92 - 30794189 -----------CGCUCUGUAUUUGCUCUCGAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGCA------ -----------.((.(..(((..((...((((((((((........)))))((((.(((((((((.......))))))))).))))))))))))))..).)).------ ( -20.50, z-score = -0.86, R) >droPer1.super_7 3752630 92 + 4445127 -----------CGCUCUGUAUUUGCUCUCGAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGCA------ -----------.((.(..(((..((...((((((((((........)))))((((.(((((((((.......))))))))).))))))))))))))..).)).------ ( -20.50, z-score = -0.86, R) >droVir3.scaffold_13047 4659789 95 + 19223366 ----GUGUAUCUGUAUU-UAUUUGCGCUCAAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCAAUUGUUUGGCGUGUGGU--------- ----(((((..((....-))..)))))....((((...((((..(((((((.....(((((((((.......))))))))))))))))..))))..))))--------- ( -21.90, z-score = -1.83, R) >droGri2.scaffold_14830 5406059 95 - 6267026 ----GUGUGUGUGUAUU-UAUUUGCGCUCAAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGU--------- ----...((.(((((..-....))))).)).((((...((((..(((((((.....(((((((((.......))))))))))))))))..))))..))))--------- ( -23.90, z-score = -2.23, R) >consensus ____UUGUGCGAGUACU_UAUUUGCUCUCAAACCAAUUUGUUUUAUAAUUGAAUUUGAAAUUGAAGAUGACAUUCGGUUUCCGAUUGUUUGGCGUGUGGUGCU______ ...............................((((...((((..(((((((.....(((((((((.......))))))))))))))))..))))..))))......... (-18.24 = -18.16 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:27 2011