
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,343,514 – 17,343,610 |
| Length | 96 |
| Max. P | 0.869201 |

| Location | 17,343,514 – 17,343,610 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.90 |
| Shannon entropy | 0.50362 |
| G+C content | 0.42228 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -12.58 |
| Energy contribution | -12.63 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
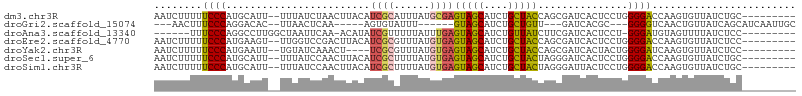
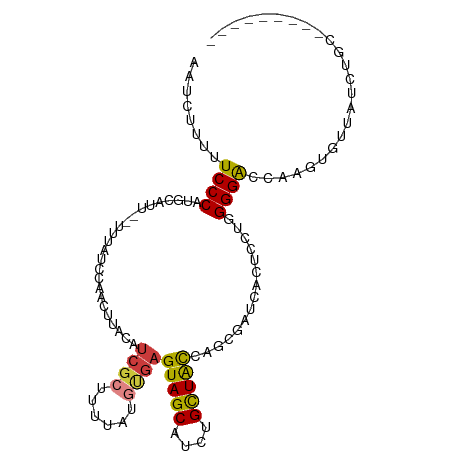
>dm3.chr3R 17343514 96 - 27905053 AAUCUUUUUCCCAUGCAUU--UUUAUCUAACUUACAUCGCAUUUAUGCGAGUAGCAUCUGCUACCAGCGAUCACUCCUGGGGACCAAGUGUUAUCUGC--------- ...(((..((((.......--...............(((((....)))))(((((....)))))(((.((....)))))))))..)))..........--------- ( -21.10, z-score = -1.53, R) >droGri2.scaffold_15074 2860576 85 - 7742996 ---AACUUUCCCAGGACAC--UUAACUCAA-----AGUGUAUUU------GUAGCAUCUGCUGUU---GAUCACGC---GGGGUCAACUGUUAUCAGCAUCAAUUGC ---........((((((((--((......)-----))))).)))------)..(((..((.((((---(((...((---((......)))).))))))).))..))) ( -21.10, z-score = -1.30, R) >droAna3.scaffold_13340 17683850 90 - 23697760 ------UUUCCCAGGCCUUGGCUAAUUCAA-ACAUAUCGUUUUUAUUUGAGUAGCAUCUGUUAUCUUCGAUCACUCCU-GGGAUGUAGUUUUAUCUCC--------- ------..(((((((.....((((.(((((-(.............))))))))))....((.(((...))).)).)))-))))...............--------- ( -18.52, z-score = -1.48, R) >droEre2.scaffold_4770 13423556 96 + 17746568 AAUCUUUUUCCCAUGAAGU--UUGGUCCGACUUACAUCGCGUUUAUGUGAGUAGCAUCUGCUACCAGCGAUCACUCCUGGGGACCAAGUGUUAUCUCC--------- ............((((..(--(((((((.((((((((.......))))))))(((....))).((((.((....))))))))))))))..))))....--------- ( -28.60, z-score = -2.66, R) >droYak2.chr3R 6173482 92 - 28832112 AAUCUUUUUCCCAUGAAUU--UGUAUCAAACU----UCGCGUUUAUGUGAGUAGCAUCUGCUACCAGCGAUCACUACUGGGGAUCAAGUGUUAUCUCC--------- ...(((..((((.......--((.(((.....----(((((....)))))(((((....)))))....)))))......))))..)))..........--------- ( -18.82, z-score = -0.70, R) >droSec1.super_6 212732 96 + 4358794 AAUCUUUUUCCCAUGCAUU--UUUAUCCAACUUACAUCGCUUUUAUGUGAGUAGCAUCUGCUACUAGGGAUCACUCCUGGGGACCAAGUGUUAUCUGC--------- ..............(((((--(...(((.((((((((.......))))))))(((....))).((((((.....)))))))))..)))))).......--------- ( -21.40, z-score = -1.06, R) >droSim1.chr3R 23392043 96 + 27517382 AAUCUUUUUCCCAUGCAUU--UUUAUCCAACUUACAUCGCUUUUAUGUGAGUAGCAUCUGCUACUAGGGAUUACUCCUGGGGACCAAGUGUUAUCUGC--------- ..............(((((--(...(((.((((((((.......))))))))(((....))).((((((.....)))))))))..)))))).......--------- ( -21.40, z-score = -1.13, R) >consensus AAUCUUUUUCCCAUGCAUU__UUUAUCCAACUUACAUCGCUUUUAUGUGAGUAGCAUCUGCUACCAGCGAUCACUCCUGGGGACCAAGUGUUAUCUGC_________ ........((((........................((((......))))(((((....)))))...............))))........................ (-12.58 = -12.63 + 0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:23 2011