| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,313,560 – 17,313,716 |
| Length | 156 |
| Max. P | 0.991206 |

| Location | 17,313,560 – 17,313,716 |
|---|---|
| Length | 156 |
| Sequences | 6 |
| Columns | 168 |
| Reading direction | reverse |
| Mean pairwise identity | 65.32 |
| Shannon entropy | 0.65292 |
| G+C content | 0.41707 |
| Mean single sequence MFE | -43.58 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.07 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
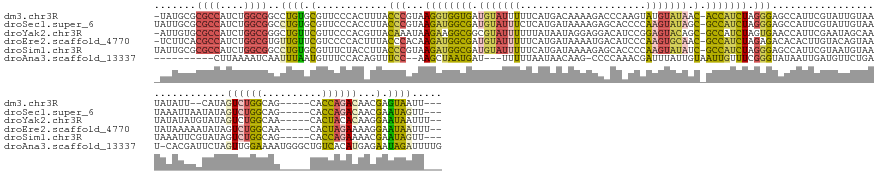
>dm3.chr3R 17313560 156 - 27905053 -UAUGCGCGCCAUCUGGCGGCCUGUGCGUUCCCACUUUACCCGUAAGGUGGUGAUGUAUUUUUCAUGACAAAAGACCCAAGUAUGUAUAAC-ACCAUCUAGGGAGCCAUUCGUAUUGUAAUAUAUU--CAUAGUCUGGCAG-----CACCAGACAACGAGUAAUU--- -.(((((.(((....)))(((..((((((..((((((((....))))))))..))))))................(((.((.(((......-..))))).))).)))...))))).......((((--(...((((((...-----..))))))...)))))...--- ( -47.80, z-score = -2.34, R) >droSec1.super_6 183265 159 + 4358794 UAUUGCGCGCCAUCUGGCGGCCUGUGCGUUCCCACCUUACCCGUAAGAUGGCGAUGUAUUUCUCAUGAUAAAAGAGCACCCCAAGUAUAGC-GCCAUCUAGGGAGCCAUUCGUAUUGUAAUAAAUUAAUAUAGUCUGGCAG-----CACCAGACAACGAAUAGUU--- ((((((.((((....))))((..(((.((((((((.......)).((((((((.((((((((((.........)))......))))))).)-))))))).)))))))))..))...))))))..........((((((...-----..))))))...........--- ( -49.80, z-score = -2.77, R) >droYak2.chr3R 6148918 159 - 28832112 -AUUGUGCGCCAUCUGGCGGGCUGUUCGUUCCCACGUUACAAAUAAGAAGGCGGCGUAUUUUUUAUAAUAGGAGGACAUCCGGAGUACAGC-GCCAUCUAGUGAACCAUUCGAAUAGCAAUAUAUAUGUAUAGUCUGGCAA-----CACUACACAAGGAAUAAUUU-- -.(((((((((....)))).((((((((.......(((.((....(((.((((..(((((((........(((.....))))))))))..)-))).)))..)))))....))))))))............((((..(....-----))))))))))..........-- ( -39.30, z-score = -0.24, R) >droEre2.scaffold_4770 13394272 159 + 17746568 -UCUUCACGCCAUCUGGCGUGUUGUUCGUCCCCACUUUACCCACAAGAUGGCGAUGUAUUUUUCAUGAUAAAAUGACAUCCCAAGUGCAAC-GCCAUCUAGAGACACACUUGUACAGUAAUAUAAAAAUAUAGUCUGGCAA-----CACUAGAAAAGGAAUAAUUU-- -....((((((....))))))((((((......(((.(((.....((((((((.(((((((.((((......))))......))))))).)-))))))).(((.....)))))).)))...............(((((...-----..)))))....))))))...-- ( -40.30, z-score = -2.90, R) >droSim1.chr3R 23362141 159 + 27517382 UAUUGCGCGCCAUCUGGCGGCCUGUGCGUUUCUACCUUACCCGUAAGAUGGCGAUGUAUUUUUCAUGAUAAAAGAGCACCCCAAGUAUAUC-GCCAUCUAGGGAGCCAUUCGUAAUGUAAUAAAUUCGUAUAGUCUGGCAG-----CACCAGAAAACGAAUAGUU--- (((((((.(((....)))(((..(((......)))....(((...((((((((((((((((....((.........))....)))))))))-))))))).))).)))...)))))))......((((((....(((((...-----..)))))..))))))....--- ( -51.40, z-score = -3.40, R) >droAna3.scaffold_13337 10457025 151 - 23293914 ----------CUUAAAAUCAAUUUAAUGUUUCCACAGUUUCC--AAGCUAAUGAU---UUUUUAAUAACAAG-CCCCAAACGAUUUAUUGUAAUUGUUUCGGGUAUAAUUGAUGUUCUGAU-CACGAUUCUAGUUGGAAAAUGGGCUGUCACAUGAGAAUAGAUUUUG ----------..(((((((.((((.((((..((.((.(((((--((.(((.((((---(....((((.((((-(((.((((((((......)))))))).))))....))).))))..)))-))......)))))))))).)))).....))))..)))).))))))) ( -32.90, z-score = -1.46, R) >consensus _AUUGCGCGCCAUCUGGCGGCCUGUGCGUUCCCACCUUACCCGUAAGAUGGCGAUGUAUUUUUCAUGAUAAAAGAACAACCCAAGUAUAGC_GCCAUCUAGGGAGCCAUUCGUAUUGUAAUAUAUUAAUAUAGUCUGGCAA_____CACCAGACAACGAAUAAUU___ .......((((....))))..((((.(............(((...(((((((..(((((((.....................)))))))...))))))).))).............................((((((..........))))))...).))))..... (-18.25 = -18.07 + -0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:19 2011