
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,155,637 – 8,155,739 |
| Length | 102 |
| Max. P | 0.999950 |

| Location | 8,155,637 – 8,155,739 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 56.38 |
| Shannon entropy | 0.84051 |
| G+C content | 0.31010 |
| Mean single sequence MFE | -20.88 |
| Consensus MFE | -10.36 |
| Energy contribution | -12.01 |
| Covariance contribution | 1.65 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.14 |
| SVM RNA-class probability | 0.999950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
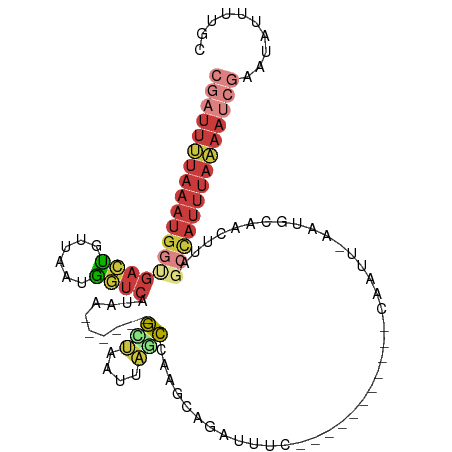
>dm3.chr2L 8155637 102 + 23011544 CGAUUUUAAAUGGUGACUGUUAAUGGUCAUAA------GCUAAUUAGCCAAGCAGCUAUCU----UACAAACAUUAAAAUGCAACUUAGCCAUUUAAAAUCGAAUAUUUUGC ((((((((((((((((((......))))))..------(((((.((((......))))...----......(((....)))....))))))))))))))))).......... ( -24.90, z-score = -3.13, R) >droSim1.chr2L 7937776 99 + 22036055 CGAUUUUAAAUGGUGACUGUUAAUGGUCAUAA------GCUAAUUAGCCAAGCUGAUUUC-------AAAACAAUUUAAUGCAACUCAGCCAUUUAAAAUCGAAUAUUUUGC ((((((((((((((((((......))))....------((.(((((((...)))))))..-------.............))......)))))))))))))).......... ( -25.49, z-score = -3.57, R) >droSec1.super_3 3652869 102 + 7220098 CGAUUUUAAAUGGUGACCGUUAAUGGUCAUAA------GCUAAUUAGCCGAGCAGCUGAUU----CCAAAACAAUUUAAUGCAACUCAGCCAUUUAAAAUCGAAUAUUUUGC (((((((((((((((((((....)))))....------...(((((((......)))))))----.......................)))))))))))))).......... ( -28.60, z-score = -4.09, R) >droYak2.chr2L 10787237 77 + 22324452 CGAUUUUAAAUGGUGACUGUUAAUGGUCAUUA------GCUAAUUACCCAAGC---UUUC--------------------------UAUCCAUUUAAAAUCGAAUAUUUUUC ((((((((((((((((((......)))))..(------(((.........)))---)...--------------------------...))))))))))))).......... ( -21.30, z-score = -5.18, R) >droEre2.scaffold_4929 17061681 88 + 26641161 CGAUUUUAAAUGCAGACUGUUAGUGGUCAAAA------AAAGCUGUAUUACAAAAAUGCA------------------AUACAACUUAGCAAUUUAAAAUCGAAUAUUUUCC (((((((((((((.((((......))))....------.(((.((((((.((....)).)------------------))))).))).)).))))))))))).......... ( -22.50, z-score = -4.41, R) >droGri2.scaffold_15126 6042109 93 + 8399593 ------UAAAUGUUAAGCUUUAAUGGUCUAAAAUAAAAAACAACGUUUAAAAAAGACACAC------------ACACAACCAAUUCUAGCUAAUCACAA-CAAAUAUUUCUC ------.........((((..(((.((((....((((........))))....))))....------------.........)))..))))........-............ ( -4.94, z-score = 0.22, R) >triCas2.ChLG8 279110 112 + 15773733 CAGUUCUAAAGGGCGAAUUCAAGAAUGCAAUAAAAAUAGGUGAUUACCCUUACAAAUUUCAAAGUUGGCGCCAAUUACUCGUAGUUCAGCCAUCUUGAAAAUAAUUUUUUGU ..((((....))))...(((((((.........((((..((((......))))..))))......((((...(((((....)))))..)))))))))))............. ( -18.40, z-score = -0.00, R) >consensus CGAUUUUAAAUGGUGACUGUUAAUGGUCAUAA______GCUAAUUAGCCAAGCAGAUUUC___________CAAUU_AAUGCAACUUAGCCAUUUAAAAUCGAAUAUUUUGC ((((((((((((((((((......))))..........(((....)))........................................)))))))))))))).......... (-10.36 = -12.01 + 1.65)
| Location | 8,155,637 – 8,155,739 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 56.38 |
| Shannon entropy | 0.84051 |
| G+C content | 0.31010 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -6.93 |
| Energy contribution | -8.16 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
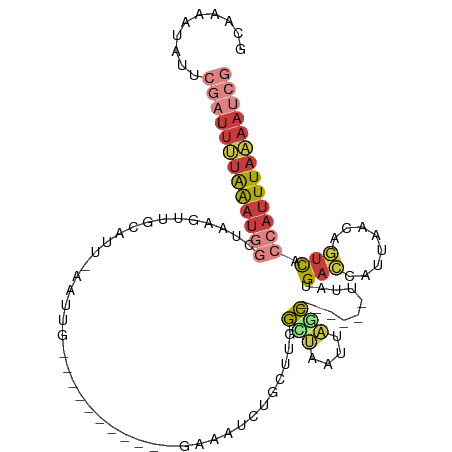
>dm3.chr2L 8155637 102 - 23011544 GCAAAAUAUUCGAUUUUAAAUGGCUAAGUUGCAUUUUAAUGUUUGUA----AGAUAGCUGCUUGGCUAAUUAGC------UUAUGACCAUUAACAGUCACCAUUUAAAAUCG ..........(((((((((((((((((((.((((((((.......))----)))).)).)))))))........------...((((........)))).)))))))))))) ( -27.80, z-score = -3.73, R) >droSim1.chr2L 7937776 99 - 22036055 GCAAAAUAUUCGAUUUUAAAUGGCUGAGUUGCAUUAAAUUGUUUU-------GAAAUCAGCUUGGCUAAUUAGC------UUAUGACCAUUAACAGUCACCAUUUAAAAUCG ..........((((((((((((((..(((((..(((((....)))-------))...)))))..))........------...((((........)))).)))))))))))) ( -26.40, z-score = -3.87, R) >droSec1.super_3 3652869 102 - 7220098 GCAAAAUAUUCGAUUUUAAAUGGCUGAGUUGCAUUAAAUUGUUUUGG----AAUCAGCUGCUCGGCUAAUUAGC------UUAUGACCAUUAACGGUCACCAUUUAAAAUCG ..........(((((((((((((((((((.((...............----.....)).)))))))........------...(((((......))))).)))))))))))) ( -30.05, z-score = -4.11, R) >droYak2.chr2L 10787237 77 - 22324452 GAAAAAUAUUCGAUUUUAAAUGGAUA--------------------------GAAA---GCUUGGGUAAUUAGC------UAAUGACCAUUAACAGUCACCAUUUAAAAUCG ..........(((((((((((((...--------------------------...(---(((.........)))------)..((((........))))))))))))))))) ( -20.20, z-score = -4.66, R) >droEre2.scaffold_4929 17061681 88 - 26641161 GGAAAAUAUUCGAUUUUAAAUUGCUAAGUUGUAU------------------UGCAUUUUUGUAAUACAGCUUU------UUUUGACCACUAACAGUCUGCAUUUAAAAUCG ..........(((((((((((.((.(((((((((------------------((((....))))))))))))).------....(((........))).))))))))))))) ( -27.00, z-score = -5.87, R) >droGri2.scaffold_15126 6042109 93 - 8399593 GAGAAAUAUUUG-UUGUGAUUAGCUAGAAUUGGUUGUGU------------GUGUGUCUUUUUUAAACGUUGUUUUUUAUUUUAGACCAUUAAAGCUUAACAUUUA------ ......((..((-(((.....(((((....)))))((..------------(((.((((....((((........))))....)))))))....)).)))))..))------ ( -9.40, z-score = 1.66, R) >triCas2.ChLG8 279110 112 - 15773733 ACAAAAAAUUAUUUUCAAGAUGGCUGAACUACGAGUAAUUGGCGCCAACUUUGAAAUUUGUAAGGGUAAUCACCUAUUUUUAUUGCAUUCUUGAAUUCGCCCUUUAGAACUG .............(((((((((((.(.....(((....))).)))))...........(((((((((....)))).......))))).)))))))(((........)))... ( -16.41, z-score = 0.56, R) >consensus GCAAAAUAUUCGAUUUUAAAUGGCUAAGUUGCAUU_AAUUG___________GAAAUCUGCUUGGCUAAUUAGC______UUAUGACCAUUAACAGUCACCAUUUAAAAUCG ..........(((((((((((((.........................................(((....)))..........(((........))).))))))))))))) ( -6.93 = -8.16 + 1.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:24 2011