
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,282,463 – 17,282,588 |
| Length | 125 |
| Max. P | 0.846025 |

| Location | 17,282,463 – 17,282,565 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Shannon entropy | 0.38501 |
| G+C content | 0.46389 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -13.63 |
| Energy contribution | -14.66 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17282463 102 + 27905053 GACUGGCCAAUGGCCAGGCAAUGUGAUUAAGCCACAGAUACACAGAUAUCUACGAGAUACAAGACACACACUCGUUCGAAUACAUGUUCAUACACAACACAC ..((((((...))))))....((((..........(((((......)))))(((((..............)))))..((((....))))...))))...... ( -20.74, z-score = -1.95, R) >droSim1.chr3R 23329854 102 - 27517382 GACUGGCCAAUGGCCAGGCAAUGUGAUUAAGCCACAGAUACACAGAUAUCUACGAGAUACAAGACACACACUCGUUCGAAUACAUGUUCAUACACACCACAC ..((((((...))))))....((((..........(((((......)))))(((((..............)))))..((((....))))...))))...... ( -20.44, z-score = -1.71, R) >droSec1.super_6 152272 102 - 4358794 GACUGGCCAAUGGCCAGGCAAUGUGAUUAAGCCACAGAUACACAGAUAUCUACGAGAUACAAGACACACACUCGUUCGAAUACAUGUUCAUACACACCACAC ..((((((...))))))....((((..........(((((......)))))(((((..............)))))..((((....))))...))))...... ( -20.44, z-score = -1.71, R) >droYak2.chr3R 6114270 99 + 28832112 GACUGGCCAAUGGCCAGGCAAUGUGAUUAAGCCACAGAUACACAGAUAUCUACGAGAUACGAGACACACACCCGUUCGAAUGCAUGUACAUAUACUUCC--- ..((((((...))))))(((..(((.......)))(((((......))))).(((...(((.(........)))))))..)))................--- ( -19.00, z-score = -0.54, R) >droEre2.scaffold_4770 13361798 93 - 17746568 GACUGGCCAAUGGCCAGGCAAUGUGAUUAAGCCACAGAUACACAGAUAUCUACGAGAUACAAGACACACACUCGUUCGAAUGCAUGUAUAUUU--------- ..((((((...))))))(((.(((((((........))).))))....((.(((((..............)))))..)).)))..........--------- ( -20.24, z-score = -1.35, R) >droAna3.scaffold_13340 17625554 88 + 23697760 GCCUGGCCAAUGGCCAAGCAAUGUGAUUAAC------ACACACAAUCUCCUGGAAGCCCAGACACUCACGCGCACACAGACACAUGGCCCCAUA-------- ((((((((...))))).((..((((.....)------))).........((((....))))........))..............)))......-------- ( -20.60, z-score = -0.74, R) >consensus GACUGGCCAAUGGCCAGGCAAUGUGAUUAAGCCACAGAUACACAGAUAUCUACGAGAUACAAGACACACACUCGUUCGAAUACAUGUUCAUACACA_CA___ ..((((((...))))))...(((((..........(((((......)))))(((((..............))))).....)))))................. (-13.63 = -14.66 + 1.03)



| Location | 17,282,463 – 17,282,565 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.68 |
| Shannon entropy | 0.38501 |
| G+C content | 0.46389 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.846025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17282463 102 - 27905053 GUGUGUUGUGUAUGAACAUGUAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUGGCUUAAUCACAUUGCCUGGCCAUUGGCCAGUC (((((....(((..((((..(((((....)))))..))).)..))).((((((((((....))))))))))......)))))...((((((...)))))).. ( -28.60, z-score = -1.48, R) >droSim1.chr3R 23329854 102 + 27517382 GUGUGGUGUGUAUGAACAUGUAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUGGCUUAAUCACAUUGCCUGGCCAUUGGCCAGUC ..(..(((((((..((((..(((((....)))))..))).)..))..((((((((((....))))))))))......)))))..)((((((...)))))).. ( -32.80, z-score = -2.50, R) >droSec1.super_6 152272 102 + 4358794 GUGUGGUGUGUAUGAACAUGUAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUGGCUUAAUCACAUUGCCUGGCCAUUGGCCAGUC ..(..(((((((..((((..(((((....)))))..))).)..))..((((((((((....))))))))))......)))))..)((((((...)))))).. ( -32.80, z-score = -2.50, R) >droYak2.chr3R 6114270 99 - 28832112 ---GGAAGUAUAUGUACAUGCAUUCGAACGGGUGUGUGUCUCGUAUCUCGUAGAUAUCUGUGUAUCUGUGGCUUAAUCACAUUGCCUGGCCAUUGGCCAGUC ---..(((((((((.((((((((((....))))))))))..)))))..(((((((((....)))))))))))))...........((((((...)))))).. ( -29.40, z-score = -1.20, R) >droEre2.scaffold_4770 13361798 93 + 17746568 ---------AAAUAUACAUGCAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUGGCUUAAUCACAUUGCCUGGCCAUUGGCCAGUC ---------..(((.((((((((((....))))))))))..)))...((((((((((....))))))))))..............((((((...)))))).. ( -25.70, z-score = -1.56, R) >droAna3.scaffold_13340 17625554 88 - 23697760 --------UAUGGGGCCAUGUGUCUGUGUGCGCGUGAGUGUCUGGGCUUCCAGGAGAUUGUGUGU------GUUAAUCACAUUGCUUGGCCAUUGGCCAGGC --------.........(((((.....(..((((..(...(((((....)))))...)..)))).------.)....))))).((((((((...)))))))) ( -32.40, z-score = -1.07, R) >consensus ___UG_UGUAUAUGAACAUGUAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUGGCUUAAUCACAUUGCCUGGCCAUUGGCCAGUC .........(((((((((..(((((....)))))..))).))))))..(((((((((....)))))))))...............((((((...)))))).. (-19.59 = -19.93 + 0.34)

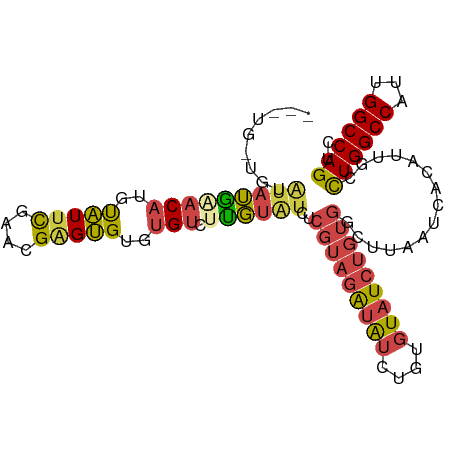

| Location | 17,282,495 – 17,282,588 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 75.52 |
| Shannon entropy | 0.46434 |
| G+C content | 0.46732 |
| Mean single sequence MFE | -21.81 |
| Consensus MFE | -12.61 |
| Energy contribution | -12.95 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17282495 93 - 27905053 CUUCGCAGUGUCCCUGUGUGUAUGUGUGUUGUGUAUGAACAUGUAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUG ...(((((((..(..((((.((((.(......(((..((((..(((((....)))))..))).)..)))).)))).))))..)..)).))))) ( -20.50, z-score = -1.56, R) >droSim1.chr3R 23329886 93 + 27517382 CUUCGCAGUGUCCCUGUGUGUGUGUGUGGUGUGUAUGAACAUGUAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUG ...(((((.....))))).(((..((.(((((.((((((((..(((((....)))))..)))........))))).)))))))..)))..... ( -21.20, z-score = -1.47, R) >droSec1.super_6 152304 93 + 4358794 CUUCGCAGUAUCCCUGUGUGUGUGUGUGGUGUGUAUGAACAUGUAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUG ...(((((.....))))).(((..((.(((((.((((((((..(((((....)))))..)))........))))).)))))))..)))..... ( -21.20, z-score = -1.36, R) >droYak2.chr3R 6114302 90 - 28832112 CUUCGCAGUGCCCCUGUGUGUGUG---GAAGUAUAUGUACAUGCAUUCGAACGGGUGUGUGUCUCGUAUCUCGUAGAUAUCUGUGUAUCUGUG ..(((.(((((...((((..((((---....))))..)))).))))))))(((((((..(((((((.....)).))))....)..))))))). ( -23.90, z-score = -0.92, R) >droEre2.scaffold_4770 13361830 84 + 17746568 CUUCGCAGUGUCCCUGUGUGUGU---------AAAUAUACAUGCAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUG ...(((((((((..(((((((((---------....))))))))).....(((((..............))))).)))).)))))........ ( -20.14, z-score = -1.88, R) >droAna3.scaffold_13340 17625583 80 - 23697760 CUUCGGAGACUCCGAGCGUCU----------UAUGGGGCCAUGUGUCUGUGUGCGCGUGAGUGUCUGGGCUUCCAGGAGAUUGUGUGUGU--- ...((.((((.(...((.((.----------...)).))...).)))).)).(((((..(...(((((....)))))...)..)))))..--- ( -23.90, z-score = 0.22, R) >consensus CUUCGCAGUGUCCCUGUGUGUGUG___G_UGUAUAUGAACAUGUAUUCGAACGAGUGUGUGUCUUGUAUCUCGUAGAUAUCUGUGUAUCUGUG ...(((((.....)))))..............(((((((((..(((((....)))))..))).))))))....(((((((....))))))).. (-12.61 = -12.95 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:12 2011