
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,282,092 – 17,282,184 |
| Length | 92 |
| Max. P | 0.627096 |

| Location | 17,282,092 – 17,282,184 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.94 |
| Shannon entropy | 0.48575 |
| G+C content | 0.54596 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -18.32 |
| Energy contribution | -18.61 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17282092 92 - 27905053 --UCACAUUCCCAUGACAU-UCUGCAAGAUCGCUCAGCUCAGCAA-AACGCCAGCGGAAGUGGCAAUAUGAGUGGCGUUUCAAGACGCCGCCACGA------------ --(((.(((.((((....(-(((((..((.(.....).)).((..-...))..)))))))))).))).)))((((((((....)))))))).....------------ ( -26.30, z-score = -0.82, R) >droSim1.chr3R 23329471 92 + 27517382 --UCACAUUCCCAUGACAU-UCUGCAAGAUCGCUCAGCUCAGCAA-AACGCCAGCGGAAGUGGCAAUAUGAGUGGCGUUUCAAGACGCCGCCACGA------------ --(((.(((.((((....(-(((((..((.(.....).)).((..-...))..)))))))))).))).)))((((((((....)))))))).....------------ ( -26.30, z-score = -0.82, R) >droSec1.super_6 151888 92 + 4358794 --UCACAUUCCCAUGACAU-UCUGCAAGAUCGCUCAGCUCAGCAA-AACGCCAGCGGAAGUGGCAAUAUGAGUGGCGUUUCAAGACGCCGCCACGA------------ --(((.(((.((((....(-(((((..((.(.....).)).((..-...))..)))))))))).))).)))((((((((....)))))))).....------------ ( -26.30, z-score = -0.82, R) >droYak2.chr3R 6113887 92 - 28832112 --UCACAUUCCCAUGACAU-UCUGCAAGAUCGCUCAGCUCAGCAA-AACGCCAGCGGAAGUGGCAAUAUGAGUGGCGUUUCAAGACGCCGGCACGA------------ --.................-(((((......(((((((...))..-...((((.(....)))))....)))))((((((....)))))).))).))------------ ( -26.30, z-score = -0.52, R) >droEre2.scaffold_4770 13361430 92 + 17746568 --UCACAUUCCCAUGACAU-UCUGCAAGAUCGCUCAGCUCAGCAA-AACGCCAGCGGAAGUGGCAAUAUGAGUGGCGUUUCAAGACGCCGCCACGA------------ --(((.(((.((((....(-(((((..((.(.....).)).((..-...))..)))))))))).))).)))((((((((....)))))))).....------------ ( -26.30, z-score = -0.82, R) >dp4.chr2 10720232 88 - 30794189 -UUCACAUUCCCAUGACAU-UCUGCAGA---CCUCGAACGGAGGA---CGCCAGCGGAAGUGGCAAAAUGAGUGGCGUUUCAAGACGCCGCCAAGC------------ -..................-...((...---((((.....)))).---.((((.(....)))))....((.((((((((....)))))))))).))------------ ( -28.20, z-score = -1.46, R) >droPer1.super_0 2064258 88 + 11822988 -UUCACAUUCCCAUGACAU-UCUGCAGG---CCUCGAACGGAGGA---CGCCAGCGGAAGUGGCAAAAUGAGUGGCGUUUCAAGACGCCGCCAAGC------------ -....((((.((((....(-(((((.((---((((.....)))..---.))).))))))))))...)))).((((((((....)))))))).....------------ ( -30.10, z-score = -1.50, R) >droWil1.scaffold_181089 8149796 97 - 12369635 --UCACAUUCCCAUGACAU-UUUGCCAU---CCCCAAAGAGAU-----CGACAACGGAAGUGGCAAUAUGAGUGGCGUUUCAAGACGCCGCCGCCGCCGCCACGACAA --..............(((-.(((((((---..((........-----.......))..))))))).))).((((((((....))))))))................. ( -25.46, z-score = -1.26, R) >droVir3.scaffold_12822 3076974 80 + 4096053 ---------CAGACGAGUAAUUCCCAUG-----ACAGCCAGCUA--CGAUCCGGCGGAAGUGGCAAAUCGAGUGGCGUUUCAAGACGCCGCCCGGA------------ ---------...................-----.......((((--(..(((...))).)))))...(((.((((((((....)))))))).))).------------ ( -24.50, z-score = -0.62, R) >droGri2.scaffold_15074 2797738 93 - 7742996 ACUCGUAUUCCCAUGACAUCUCUGUAGG---GCCCAAACGAUCGAUCUCCUCAGCGGAAGUGGCAAACUGUAUGGCGUUUCAAGACGCCGCCGACA------------ ..((((...(((...(((....))).))---).....))))((..........((....))(((.........((((((....)))))))))))..------------ ( -20.40, z-score = 1.17, R) >consensus __UCACAUUCCCAUGACAU_UCUGCAAG___GCUCAGCUCAGCAA_AACGCCAGCGGAAGUGGCAAUAUGAGUGGCGUUUCAAGACGCCGCCACGA____________ .................................................((((.(....)))))....((.((((((((....))))))))))............... (-18.32 = -18.61 + 0.29)
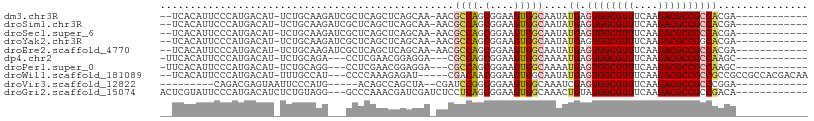

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:10 2011