
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,264,531 – 17,264,636 |
| Length | 105 |
| Max. P | 0.762421 |

| Location | 17,264,531 – 17,264,636 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 69.88 |
| Shannon entropy | 0.64416 |
| G+C content | 0.48293 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -11.55 |
| Energy contribution | -11.22 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.762421 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 17264531 105 + 27905053 CCUCGGCUCCAUUGCUCGAUCGGCUGUUGUUGGGCUUCUCCAAACUGGUUCUGGGCCCGCU--CUCUCACAACU-UAGUUGUAA--UUGCAGUCCUUUGAUUGCAUUUGU ..(((((......)).))).(((((((((((((((((..((.....))....)))))))..--.....))))).-.)))))(((--.(((((((....))))))).))). ( -28.91, z-score = -1.31, R) >droSim1.chr3R 23311829 105 - 27517382 CCUCGGCUCCAUUGCUCGAUCGGCUGUUGUUGGACUUCUCCAAACUGGUUCUGGGCCCGCU--CUCUCGCAACU-UAGUUGUAA--UUGCAGUCCUUUGAUUGCAUUUGU ..(((((......)).))).((((((((((.(((.....((.....))....(((....))--)))).))))).-.)))))(((--.(((((((....))))))).))). ( -24.40, z-score = 0.10, R) >droSec1.super_6 134432 105 - 4358794 CCUCGGCUCCAUUGCUCGAUCGGCUGUUGUUGGGCUUCUCCAAACUGGUUCUGGGCCCGCU--CUCUCGCAACU-UAGUUGUAA--UUGCAGUCCUUUGAUUGCAUUUGU ..(((((......)).))).((((((..(((((((((..((.....))....))))))((.--.....))))).-))))))(((--.(((((((....))))))).))). ( -30.10, z-score = -1.42, R) >droYak2.chr3R 6095164 102 + 28832112 CCUCGGUUCCAUUGCUCGGUCGGCUGUUU---GGCUUCUCCAAACUGGUUCUGGGCCCGCU--CUCUCACAACU-UAGUUGUAA--UUGCAGUCCUUUGAUUGCAUUUGU .(((((..(((..(((.....))).((((---((.....)))))))))..)))))......--.....(((((.-..)))))..--.(((((((....)))))))..... ( -25.70, z-score = -0.94, R) >droEre2.scaffold_4770 13343500 102 - 17746568 CCUCGGUUCCAUUGCGCGCUCGGCUGUUU---GGCUUCUCCAAACUGGUUCUGGGCCCGCU--CUCUCACAACU-UAGUUGUAA--UUGCAGUCCUUUGAUUGCAUUUGU .............(((.((((((..((((---((.....)))))).....)))))).))).--.....(((((.-..)))))..--.(((((((....)))))))..... ( -28.00, z-score = -1.45, R) >droAna3.scaffold_13340 17606424 100 + 23697760 ACACAAUUCUCUUCCAUGGCCUCUCCGAGCCGAACU---CCGAACUGGUU-UGGGCCCAG---CUCCGGCAACU-AAGUUGUAA--UUGCAGUCCUUUGAUUGCAUUUGU .................((((....(((((((....---......)))))-))))))(((---((..(....).-.)))))(((--.(((((((....))))))).))). ( -25.80, z-score = -0.76, R) >dp4.chr2 10702134 108 + 30794189 UCUGGGUUUCGCU-CGCGCUCUCCCUCUCUCUCGCUGGCCCUCUCUCUUAACUGGUUUGUUCCCUCUCACAACU-AAGUUGUAAAAUUGCAGUCCUUUGAUUGCAUUUGU ...(((....(((-.(((..............))).)))...)))...(((((((((((........)).))))-.)))))......(((((((....)))))))..... ( -18.14, z-score = -0.55, R) >droPer1.super_0 2045929 90 - 11822988 -------------------UCUGGGUUUCGCUCGCUGGCCCUCUCUCUUAACUGGUUUGUUCCCUCUCACAACU-AAGUUGUAAAAUUGCAGUCCUUUGAUUGCAUUUGU -------------------...(((((.........))))).......(((((((((((........)).))))-.)))))......(((((((....)))))))..... ( -16.70, z-score = -0.59, R) >droWil1.scaffold_181089 8128817 106 + 12369635 CGUCAGUUCUCUUAACUGGUUUUCACAUGUC--GUUUCUCUCAAUUCUCUCAGCUUUCUGUCUCUCUUGUAAGUCAAAUUGCAA--UUGCAGUCCUUUGAUUGCAUUUGU ..((((((.....))))))............--....((..(((......(((....)))......)))..)).......((((--.(((((((....))))))).)))) ( -16.20, z-score = -1.38, R) >consensus CCUCGGUUCCAUUGCUCGAUCGGCUGUUGUCGGGCUUCUCCAAACUGGUUCUGGGCCCGCU__CUCUCACAACU_UAGUUGUAA__UUGCAGUCCUUUGAUUGCAUUUGU ....................................................................(((((....))))).....(((((((....)))))))..... (-11.55 = -11.22 + -0.33)

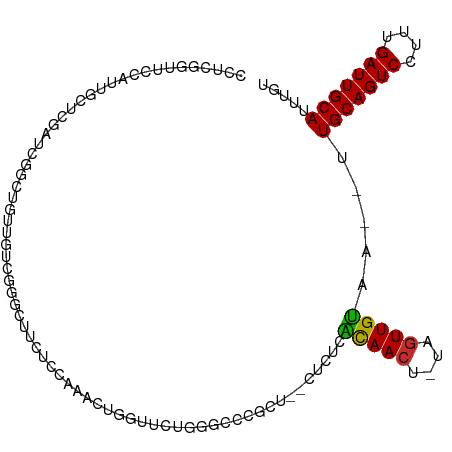
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:08 2011