| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,216,968 – 17,217,078 |
| Length | 110 |
| Max. P | 0.998180 |

| Location | 17,216,968 – 17,217,078 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.80 |
| Shannon entropy | 0.55817 |
| G+C content | 0.53165 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -17.82 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
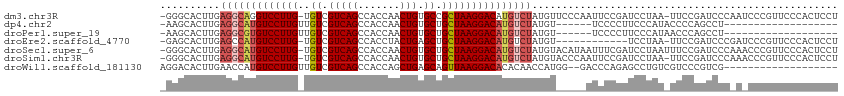
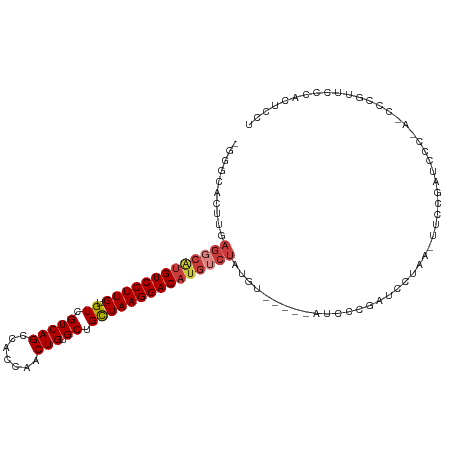
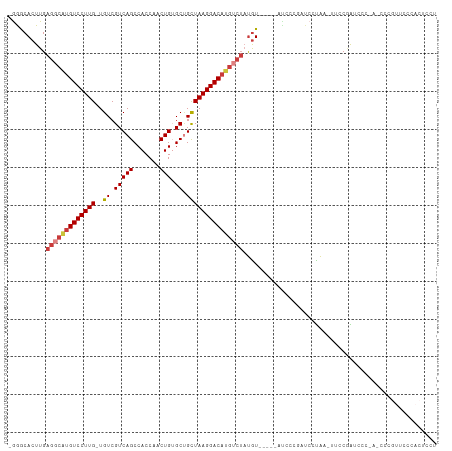
>dm3.chr3R 17216968 110 - 27905053 -GGGCACUUGAGGCAGGUCCUUG-UGUCGUCAGCCACCAACUGUGCCGCUAAGGACAUGUCUAUGUUCCCAAUUCCGAUCCUAA-UUCCGAUCCCAAUCCCGUUCCCACUCCU -(((.((...(((((.(((((((-((.((.(((.......))))).))).)))))).)))))..)).)))......((((....-....)))).................... ( -29.80, z-score = -1.96, R) >dp4.chr2 5715741 87 - 30794189 -AAGCACUUGAGGCAUGUCCUUGUUGUCGUCAGCCACCAACUGUGCUGCUAAGGACAUGUCUAUGU------UCCCCUUCCCAUACCCCAGCCU------------------- -.((((....(((((((((((((.....(.((((((.....)).)))))))))))))))))).)))------).....................------------------- ( -23.70, z-score = -2.20, R) >droPer1.super_19 1444451 87 - 1869541 -AAGCACUUGAGGCGUGUCCUUGUUGUCGUCAGCCACCAACUGUGCUGCUAAGGACAUGUCUAUGU------UCCCCUUCCCAUAACCCAGCCU------------------- -.((((....(((((((((((((.....(.((((((.....)).)))))))))))))))))).)))------).....................------------------- ( -23.30, z-score = -1.91, R) >droEre2.scaffold_4770 13294892 98 + 17746568 -GAGCACUUGAGCCAUGUCCUUG-UGUCGUCAGCCACCUACUGAGCUGCUAAGGACAUGUCUAUGU------------UCCUAA-UUCCGAUCCCGAUCCCGUUCCCACUCCU -(((((....((.((((((((((-.((.(((((.......)))).).)))))))))))).)).)))------------))....-....(((....))).............. ( -23.80, z-score = -2.17, R) >droSec1.super_6 85772 111 + 4358794 -GGGCACUUGAGGCAUGUCCUUG-UGUCGUCAGCCACCAACUGUGCUGCUAAGGACAUGUCUAUGUACAUAAUUUCGAUCCUAAUUUCCGAUCCCAAACCCGUUCCCACUCCU -(((.((...(((((((((((((-.((.(((((.......))).)).)))))))))))))))..............((((.........))))........)).)))...... ( -31.00, z-score = -2.94, R) >droSim1.chr3R 23260727 110 + 27517382 -GGGCACUUGAGGCAUGUCCUUG-UGUCGUCAGCCACCAACUGUGCUGCUAAGGACAUGUCUAUGUACCCAAUUCCGAUCCUAA-UUCCGAUCCCAAACCCGUUCCCACUCCU -(((.((...(((((((((((((-.((.(((((.......))).)).)))))))))))))))..)).)))......((((....-....)))).................... ( -33.10, z-score = -3.59, R) >droWil1.scaffold_181130 16215877 92 - 16660200 AGGACACUUGAACCAUGUCCUUGUUGUCGUCAGCCACCAGCUGAGCAGUUAAGGACACACAACCAUGG--GACCCAGAGCCUGUCGUCCCGUCG------------------- ..(((..........((((((((..((..(((((.....)))))))...)))))))).........((--(((.(((...)))..)))))))).------------------- ( -29.30, z-score = -1.32, R) >consensus _GGGCACUUGAGGCAUGUCCUUG_UGUCGUCAGCCACCAACUGUGCUGCUAAGGACAUGUCUAUGU_____AUCCCGAUCCUAA_UUCCGAUCCC_A_CCCGUUCCCACUCCU ..........(((((((((((((..((.(((((.......))).)).)))))))))))))))................................................... (-17.82 = -18.60 + 0.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:07 2011