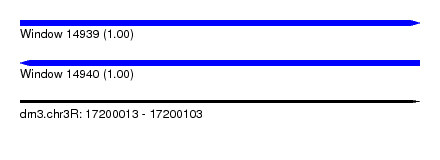
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,200,013 – 17,200,103 |
| Length | 90 |
| Max. P | 0.999949 |

| Location | 17,200,013 – 17,200,103 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.92 |
| Shannon entropy | 0.58724 |
| G+C content | 0.42011 |
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -9.88 |
| Energy contribution | -9.88 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17200013 90 + 27905053 UUUUAAGACUCGGCAAUCAAUGCCGUUGCUGGAUUUGCAU--CA-----GCAAGCUUUUACGUUGCUUAA-------ACAAAU--CCAGACAACAUCUUUAAGUAC------- ....((((...((((.....))))(((((((((((((...--.(-----((((.(......))))))...-------.)))))--)))).)))).)))).......------- ( -28.00, z-score = -4.60, R) >droAna3.scaffold_13340 215733 92 - 23697760 ACUAAAGAUUCGGCACUCAAUGCCGUUGCU-AGACAAUUGCGAU----CGCAAGCUUUUACGUUGCUUCA-------ACAAGU--CUGAACAACAUCUUUGUAUUA------- ..(((((((..((((.....))))((((.(-((((..((((...----.))))........((((...))-------))..))--)))..))))))))))).....------- ( -23.20, z-score = -2.15, R) >droEre2.scaffold_4770 13275141 90 - 17746568 UUUGAAGACUCGGCAAUUAAUGCCGUUGCUGGAUUUGCAU--CA-----GCAAGCUUUUACGUUGCUUGA-------ACAAAU--CCAGACAACAUCUUAAGGUAC------- ....((((...((((.....))))(((((((((((((...--..-----.(((((.........))))).-------.)))))--)))).)))).)))).......------- ( -29.80, z-score = -4.03, R) >droYak2.chr3R 6005945 90 + 28832112 UUUGAAGACUCGGCAAUGAAUGCCGUUGCUGGAUUUGCAU--CA-----GCAAGCUUUUACGUUGCUUGA-------ACAAAU--CCAGACAACAUCUUUAGGUAC------- ((((((((...((((.....))))(((((((((((((...--..-----.(((((.........))))).-------.)))))--)))).)))).))))))))...------- ( -32.70, z-score = -4.65, R) >droSec1.super_6 68132 90 - 4358794 UUCUAAGACUCGGCAAUCAAUGCCGUUGCUGGAUUUGCAU--CA-----GCAAGCUUUUACGUUGCUUGA-------ACAAAU--CCUGGCAACAUCUUUAAGUAC------- ....((((...((((.....))))(((((((((((((...--..-----.(((((.........))))).-------.)))))--)).)))))).)))).......------- ( -28.30, z-score = -3.52, R) >droSim1.chr3R 23242585 90 - 27517382 UUCUAAGACUCGGCAAUCAAUGCCGUUGCUGGAUUUGCAU--CA-----GCAAGCUUUUACGUUGCUUGA-------ACAAAU--CCAGACAACAUCUUUAAAUAC------- ....((((...((((.....))))(((((((((((((...--..-----.(((((.........))))).-------.)))))--)))).)))).)))).......------- ( -29.00, z-score = -4.91, R) >droWil1.scaffold_181130 16195628 112 + 16660200 -ACCAAGACUCGGCAAUAAAUGCCGUUGUUUGAACCAAAAUGGGGAAAAGCAAGCUUUUACGUUGCUACCCCUUUUCACAGGUACCAAAACAACAUCUUUAAAUUGUCCCUCA -.....(((..((((.....))))(((((((..(((((((.((((...(((((.(......)))))).))))))))....)))....)))))))...........)))..... ( -27.40, z-score = -2.13, R) >droVir3.scaffold_12822 2982847 89 - 4096053 UCCCAAGACUCGGCAAUUAAUGCCGUUGCUCGAACCA--AUGGC----AGCAAGCUUUUAUGUUGCUCCA-------AAGGGGUUCGUAACAACAUCUUUAC----------- ....((((...((((.....))))((((..((((((.--.(((.----(((((.(......)))))))))-------....))))))...)))).))))...----------- ( -28.20, z-score = -3.25, R) >droMoj3.scaffold_6540 393465 88 - 34148556 UUGUAAGACUCGGCAAUCAAUGCCGUUGCUCGAAUCACCGU-CA-----GCAAGCUUUUACGUUGCUGCG-------GCUAUU--CAAAGCAACAUCUUAUUC---------- ..((((((...((((.....))))((((((.((((..(((.-((-----((((.(......)))))))))-------)..)))--)..)))))).))))))..---------- ( -31.80, z-score = -4.98, R) >droGri2.scaffold_15074 2712922 91 + 7742996 ----AAGAUUCGGCAAUUAAUGCCGUUAUUAGAUCUCUCUGGCA-----GCAAGCUUUUAUGUUGCCCCA-------GACGGA--UCUGACAACAUCUUAACGCAUAAC---- ----(((((..((((.....))))(((.((((((((.(((((((-----(((........)))))))..)-------)).)))--))))).))))))))..........---- ( -31.50, z-score = -5.00, R) >consensus UUCUAAGACUCGGCAAUCAAUGCCGUUGCUGGAUUUGCAU__CA_____GCAAGCUUUUACGUUGCUUCA_______ACAAAU__CCAGACAACAUCUUUAAGUAC_______ ....((((..(((((.....)))))........................((((.(......))))).............................)))).............. ( -9.88 = -9.88 + -0.00)

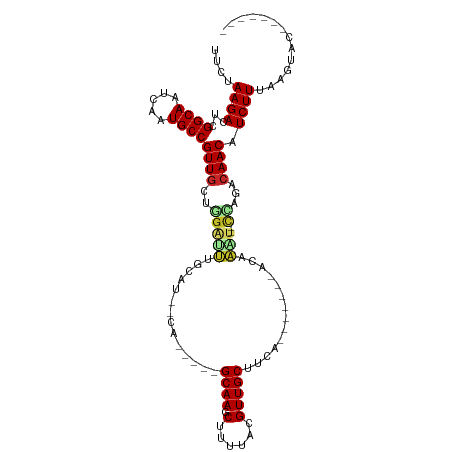
| Location | 17,200,013 – 17,200,103 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.92 |
| Shannon entropy | 0.58724 |
| G+C content | 0.42011 |
| Mean single sequence MFE | -34.19 |
| Consensus MFE | -15.91 |
| Energy contribution | -15.00 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.60 |
| Mean z-score | -4.57 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.13 |
| SVM RNA-class probability | 0.999949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17200013 90 - 27905053 -------GUACUUAAAGAUGUUGUCUGG--AUUUGU-------UUAAGCAACGUAAAAGCUUGC-----UG--AUGCAAAUCCAGCAACGGCAUUGAUUGCCGAGUCUUAAAA -------.......(((((((((.((((--((((((-------...(((((.((....))))))-----).--..))))))))))))))((((.....))))..))))).... ( -36.00, z-score = -6.07, R) >droAna3.scaffold_13340 215733 92 + 23697760 -------UAAUACAAAGAUGUUGUUCAG--ACUUGU-------UGAAGCAACGUAAAAGCUUGCG----AUCGCAAUUGUCU-AGCAACGGCAUUGAGUGCCGAAUCUUUAGU -------......((((((((((((.((--((..((-------((..((((.((....)))))).----....)))).))))-))))))(((((...)))))..))))))... ( -27.00, z-score = -2.27, R) >droEre2.scaffold_4770 13275141 90 + 17746568 -------GUACCUUAAGAUGUUGUCUGG--AUUUGU-------UCAAGCAACGUAAAAGCUUGC-----UG--AUGCAAAUCCAGCAACGGCAUUAAUUGCCGAGUCUUCAAA -------.......(((((((((.((((--((((((-------...(((((.((....))))))-----).--..))))))))))))))((((.....))))..))))).... ( -35.30, z-score = -5.73, R) >droYak2.chr3R 6005945 90 - 28832112 -------GUACCUAAAGAUGUUGUCUGG--AUUUGU-------UCAAGCAACGUAAAAGCUUGC-----UG--AUGCAAAUCCAGCAACGGCAUUCAUUGCCGAGUCUUCAAA -------.......(((((((((.((((--((((((-------...(((((.((....))))))-----).--..))))))))))))))((((.....))))..))))).... ( -35.40, z-score = -5.64, R) >droSec1.super_6 68132 90 + 4358794 -------GUACUUAAAGAUGUUGCCAGG--AUUUGU-------UCAAGCAACGUAAAAGCUUGC-----UG--AUGCAAAUCCAGCAACGGCAUUGAUUGCCGAGUCUUAGAA -------.......((((((((((..((--((((((-------...(((((.((....))))))-----).--..)))))))).)))))((((.....))))..))))).... ( -33.50, z-score = -4.54, R) >droSim1.chr3R 23242585 90 + 27517382 -------GUAUUUAAAGAUGUUGUCUGG--AUUUGU-------UCAAGCAACGUAAAAGCUUGC-----UG--AUGCAAAUCCAGCAACGGCAUUGAUUGCCGAGUCUUAGAA -------.......(((((((((.((((--((((((-------...(((((.((....))))))-----).--..))))))))))))))((((.....))))..))))).... ( -36.00, z-score = -5.64, R) >droWil1.scaffold_181130 16195628 112 - 16660200 UGAGGGACAAUUUAAAGAUGUUGUUUUGGUACCUGUGAAAAGGGGUAGCAACGUAAAAGCUUGCUUUUCCCCAUUUUGGUUCAAACAACGGCAUUUAUUGCCGAGUCUUGGU- ...(((((...........(((((((.((.(((...((((.((((.(((((.((....)))))))...)))).))))))))))))))))((((.....))))..)))))...- ( -35.40, z-score = -2.43, R) >droVir3.scaffold_12822 2982847 89 + 4096053 -----------GUAAAGAUGUUGUUACGAACCCCUU-------UGGAGCAACAUAAAAGCUUGCU----GCCAU--UGGUUCGAGCAACGGCAUUAAUUGCCGAGUCUUGGGA -----------...(((((((((((.((((((....-------(((((((((......).)))))----.))).--.))))))))))))((((.....))))..))))).... ( -33.30, z-score = -3.80, R) >droMoj3.scaffold_6540 393465 88 + 34148556 ----------GAAUAAGAUGUUGCUUUG--AAUAGC-------CGCAGCAACGUAAAAGCUUGC-----UG-ACGGUGAUUCGAGCAACGGCAUUGAUUGCCGAGUCUUACAA ----------...(((((((((((((.(--(((.((-------((((((((.((....))))))-----))-.)))).)))))))))))((((.....))))..))))))... ( -38.70, z-score = -5.97, R) >droGri2.scaffold_15074 2712922 91 - 7742996 ----GUUAUGCGUUAAGAUGUUGUCAGA--UCCGUC-------UGGGGCAACAUAAAAGCUUGC-----UGCCAGAGAGAUCUAAUAACGGCAUUAAUUGCCGAAUCUU---- ----..........((((((((((.(((--((..((-------(((((((((......).))))-----).)))))..))))).)))))((((.....))))..)))))---- ( -31.30, z-score = -3.62, R) >consensus _______GUACUUAAAGAUGUUGUCUGG__AUUUGU_______UGAAGCAACGUAAAAGCUUGC_____UG__AUGCAAAUCCAGCAACGGCAUUGAUUGCCGAGUCUUAAAA ..............((((((((((..((....((((...........(((((......).))))...........)))).))..)))))((((.....))))..))))).... (-15.91 = -15.00 + -0.91)
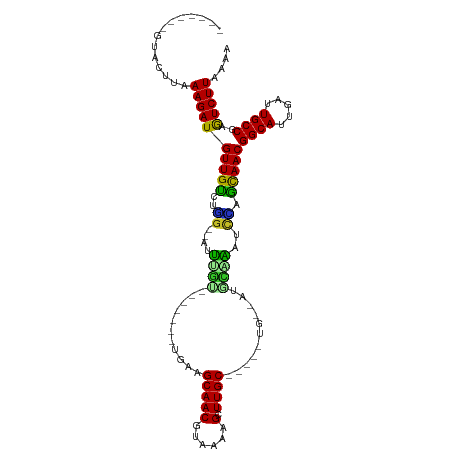
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:32:05 2011