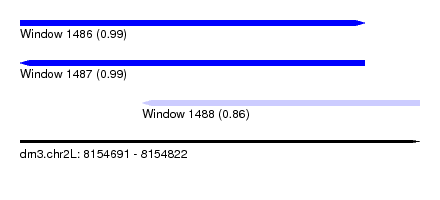
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,154,691 – 8,154,822 |
| Length | 131 |
| Max. P | 0.989199 |

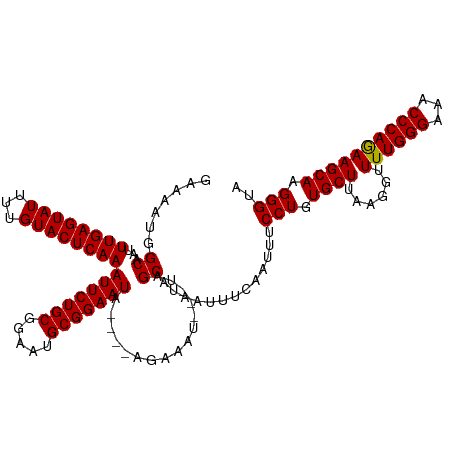
| Location | 8,154,691 – 8,154,804 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Shannon entropy | 0.14399 |
| G+C content | 0.36249 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -27.88 |
| Energy contribution | -27.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
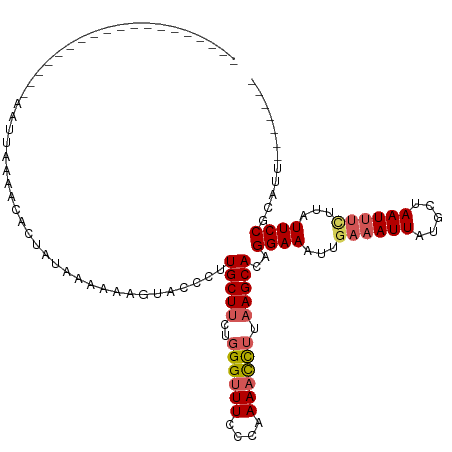
>dm3.chr2L 8154691 113 + 23011544 GAAAAUGGCAAUUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA----AGAAAU---UAGCAUAAUUUCAAUUUCCUGUGCUUAAAGUUUUGGGAAACCCAGAAGCAAGGGUA ((((...((...((((((((...))))))))(((((((.....))))))).----......---..))....)))).....(((.((((......((((((...)))))))))).))).. ( -29.80, z-score = -2.24, R) >droEre2.scaffold_4929 17060731 110 + 26641161 UAAAAUGGCAAAUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA----AUAAAU---UUGCAUAAUUUCAAUUUCCUGUGCU---GGGUUUGGGAAACCCAAAAGCAAGGGUA .......(((((((((((((...))))))..(((((((.....))))))).----...)))---)))).............(((.((((---...((.((....)).)).)))).))).. ( -32.10, z-score = -3.06, R) >droYak2.chr2L 10786317 120 + 22324452 GAAAAUGGCGAAUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUACGGAAUAAAUCAUUUGCAUAAUUUCAACUUCCUGUGCUUAAGAUUUUGGGAAACCCAGAAGCAAGGGUA ((((...(((((((((((((...))))))))(((((((.....))))))).............)))))....)))).....(((.((((......((((((...)))))))))).))).. ( -32.00, z-score = -2.31, R) >droSec1.super_3 3651937 112 + 7220098 -GAAAUGGCAAUUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA----AGAAAU---UAGCAUAAUUUCAAUUUCCUGUGCUUAAGGUUUUGGGAAACCCAGAAGCAAGGGUA -(((((.((...((((((((...))))))))(((((((.....))))))).----......---..))...))))).....(((.(((((..(((((....)))))...))))).))).. ( -33.70, z-score = -3.30, R) >droSim1.chr2L 7936842 113 + 22036055 GAAAAUGGCAAUUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUU----AGAAAU---UAGCAUAAUUUCAAUUUCCUGUGCUUAAGGUUUUGGGAAACCCAGAAGCAAGGGUA ((((...((....(((((((...)))))))((((((((.....))))))))----......---..))....)))).....(((.(((((..(((((....)))))...))))).))).. ( -31.50, z-score = -2.50, R) >consensus GAAAAUGGCAAUUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA____AGAAAU___UAGCAUAAUUUCAAUUUCCUGUGCUUAAGGUUUUGGGAAACCCAGAAGCAAGGGUA .......((...((((((((...))))))))(((((((.....)))))))................)).............(((.((((......((((((...)))))))))).))).. (-27.88 = -27.72 + -0.16)

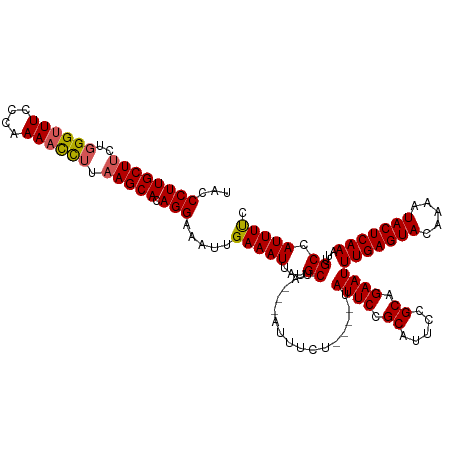
| Location | 8,154,691 – 8,154,804 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Shannon entropy | 0.14399 |
| G+C content | 0.36249 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8154691 113 - 23011544 UACCCUUGCUUCUGGGUUUCCCAAAACUUUAAGCACAGGAAAUUGAAAUUAUGCUA---AUUUCU----UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUUC ......(((((..((((((....)))))).)))))..((((...(((((((...))---))))).----..))))((.....))..((((((((((.....))))))))))......... ( -25.20, z-score = -2.94, R) >droEre2.scaffold_4929 17060731 110 - 26641161 UACCCUUGCUUUUGGGUUUCCCAAACCC---AGCACAGGAAAUUGAAAUUAUGCAA---AUUUAU----UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAUUUGCCAUUUUA ...(((((((...(((((.....)))))---)))).)))....((((((...((((---(.....----.((((.((.....)).))))(((((((.....)))))))))))).)))))) ( -25.70, z-score = -3.21, R) >droYak2.chr2L 10786317 120 - 22324452 UACCCUUGCUUCUGGGUUUCCCAAAAUCUUAAGCACAGGAAGUUGAAAUUAUGCAAAUGAUUUAUUCCGUAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAUUCGCCAUUUUC (((...(((((..((((((....)))))).)))))..((((....(((((((....))))))).)))))))....((.....)).(((((.(((((.....))))))))))......... ( -24.80, z-score = -1.94, R) >droSec1.super_3 3651937 112 - 7220098 UACCCUUGCUUCUGGGUUUCCCAAAACCUUAAGCACAGGAAAUUGAAAUUAUGCUA---AUUUCU----UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUC- ......(((((..((((((....)))))).)))))..((((...(((((((...))---))))).----..))))((.....))..((((((((((.....))))))))))........- ( -28.00, z-score = -4.08, R) >droSim1.chr2L 7936842 113 - 22036055 UACCCUUGCUUCUGGGUUUCCCAAAACCUUAAGCACAGGAAAUUGAAAUUAUGCUA---AUUUCU----AAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUUC ......(((((..((((((....)))))).)))))..((((...(((((((...))---))))).----..))))((.....))..((((((((((.....))))))))))......... ( -28.00, z-score = -3.97, R) >consensus UACCCUUGCUUCUGGGUUUCCCAAAACCUUAAGCACAGGAAAUUGAAAUUAUGCUA___AUUUCU____UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUUC ...((((((((..((((((....)))))).))))).))).....(((((...((................((((.((.....)).))))(((((((.....)))))))...)).))))). (-20.92 = -21.04 + 0.12)

| Location | 8,154,731 – 8,154,822 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Shannon entropy | 0.35814 |
| G+C content | 0.34692 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -12.48 |
| Energy contribution | -13.24 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8154731 91 - 23011544 ----------------------AAAAACACUAUAAAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACUUUAAGCACAGGAAAUUGAAAUUAUGCUAAUUUCUUAUUCCGCAUU------- ----------------------.................((.....(((((..((((((....)))))).)))))..((((...(((((((...)))))))...))))))...------- ( -15.20, z-score = -1.54, R) >droEre2.scaffold_4929 17060771 110 - 26641161 GCCCUAGACAGAAUGGACACAUUAAGACCAACAACAAAAGUACCCUUGCUUUUGGGUUUCCCAAACCC---AGCACAGGAAAUUGAAAUUAUGCAAAUUUAUUAUUCCGCAUU------- ((.........((((....))))............(((((((....)))))))(((((.....)))))---.))...((((....(((((.....)))))....)))).....------- ( -16.70, z-score = -0.01, R) >droYak2.chr2L 10786357 102 - 22324452 ------------------ACAUUAAAACCCUCUGCAAGAUUACCCUUGCUUCUGGGUUUCCCAAAAUCUUAAGCACAGGAAGUUGAAAUUAUGCAAAUGAUUUAUUCCGUAUUCCGCAUU ------------------......((((((...(((((......)))))....)))))).............((...((((....(((((((....))))))).))))(.....)))... ( -22.00, z-score = -2.17, R) >droSec1.super_3 3651976 93 - 7220098 -------------------AAUUAAAACACCAU-AAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACCUUAAGCACAGGAAAUUGAAAUUAUGCUAAUUUCUUAUUCCGCAUU------- -------------------..............-.....((.....(((((..((((((....)))))).)))))..((((...(((((((...)))))))...))))))...------- ( -18.00, z-score = -2.40, R) >droSim1.chr2L 7936882 94 - 22036055 -------------------AAUUAAAACACUAUAAAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACCUUAAGCACAGGAAAUUGAAAUUAUGCUAAUUUCUAAUUCCGCAUU------- -------------------....................((.....(((((..((((((....)))))).)))))..((((...(((((((...)))))))...))))))...------- ( -18.00, z-score = -2.56, R) >consensus ___________________AAUUAAAACACUAUAAAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACCUUAAGCACAGGAAAUUGAAAUUAUGCUAAUUUCUUAUUCCGCAUU_______ ..............................................(((((..((((((....)))))).)))))..((((...((((((.....))))))...))))............ (-12.48 = -13.24 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:22 2011