
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,123,616 – 17,123,736 |
| Length | 120 |
| Max. P | 0.999478 |

| Location | 17,123,616 – 17,123,717 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 61.26 |
| Shannon entropy | 0.74777 |
| G+C content | 0.51758 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -13.63 |
| Energy contribution | -12.52 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.80 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
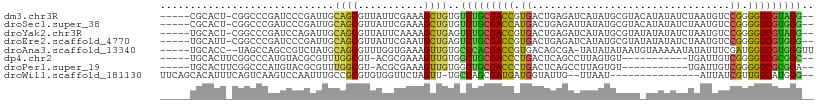
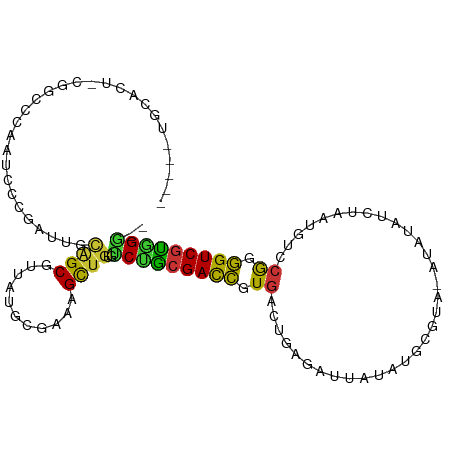
>dm3.chr3R 17123616 101 + 27905053 -----CGCACU-CGGCCCGAUCCCGAUUGCAGCGUUAUUCGAAAGCUGUGUCUGCGACCGUGACUGAGAUCAUAUGCGUACAUAUAUCUAAUGUCCGGGGUCGUAGG-- -----.(((.(-(((.......)))).)))(((...........)))....((((((((.((((..((((.(((((....)))))))))...)).)).)))))))).-- ( -33.80, z-score = -1.23, R) >droSec1.super_38 395239 101 - 400794 -----CGCACU-CGGCCCGAUCCCGAUUGCAGCGUUAUUCGAAAGCUGUGUCUGCGACCAUGACUGAGAUUAUAUGCGUACAUAUAUCUAAUGUCCGGGGUCGUGGG-- -----......-...((((((((((.((((((...(((.(....)..))).))))))....(((..((((.(((((....)))))))))...))))))))))).)).-- ( -34.00, z-score = -1.37, R) >droYak2.chr3R 5923996 101 + 28832112 -----UGCACU-CGGCCCGAUCCAGAUUGCAGCGUUAUUCAAAAGCUGAGUCUGCGACCGUGACUGAGAUCAUAUGCGUAUAUAUAUCUAAUGUCCGGGGUCGUAGG-- -----....((-((((((..((((((((.((((...........)))))))))).)).((.(((..((((.(((((....)))))))))...))))))))))).)).-- ( -34.20, z-score = -2.07, R) >droEre2.scaffold_4770 13209467 101 - 17746568 -----UGCAUU-CGGCCCGAUCCCGAUUGCAGCGUUAUUCGAAUGCUGAGUCUGCGACCGUGACUGAGAUCAUAUGCGUAUAUAUAUCUAAUGUCCGGGGUCGUGGG-- -----.(((.(-(((.......)))).)))((((((.....))))))....(..(((((.((((..((((.(((((....)))))))))...)).)).)))))..).-- ( -33.80, z-score = -1.00, R) >droAna3.scaffold_13340 3784245 101 + 23697760 -----UGCACC--UAGCCAGCCGUCUAUGCAGCGUUUGGUGAAAGUUGUGCCCACGACCGUGACAGCGA-UAUAUAUAAUGUAAAAAUAUAUUUCGAUGGUCGUGGGUU -----.(((.(--(.(((((.(((.......))).)))))...)).)))((((((((((((.....(((-.((((((.........)))))).))))))))))))))). ( -34.70, z-score = -2.99, R) >dp4.chr2 5623934 90 + 30794189 -----UGCACUUCGGCCCAUGUACGCGUUUGGCGU-ACGCGAAAGUUGUGGCUGCGACCCUGACUCAGCCUUAGUGU-----------UGAUUGUCGGGGUCGCGGC-- -----..(((..........((((((.....))))-))((....)).)))(((((((((((((((((((......))-----------)))..))))))))))))))-- ( -45.30, z-score = -4.05, R) >droPer1.super_19 1340694 90 + 1869541 -----UGCACUUCGGCCCAUGUACGCGUUUGGCGU-ACGCGAAAGUUGUGGCUGCGACCCUGACUCAGCCUUAGUGU-----------UGAUUGUCGGGGUCGCGGA-- -----........((((((.((((((.....))))-))((....)))).))))((((((((((((((((......))-----------)))..)))))))))))...-- ( -44.30, z-score = -3.87, R) >droWil1.scaffold_181130 16102510 89 + 16660200 UUCAGCACAUUUCAGUCAAGUCCAAUUUGCCGCGUGUGGUUCUAGUU-UGCCAGCGAUGAUGGUAUUG--UUAAU---------------AUUAUCGUUGUCAUGGG-- .....(((((....(.(((((...))))).)..))))).........-..((((((((((((((((..--...))---------------)))))))))))..))).-- ( -19.70, z-score = -0.45, R) >consensus _____UGCACU_CGGCCCAAUCCCGAUUGCAGCGUUAUGCGAAAGCUGUGUCUGCGACCGUGACUGAGAUUAUAUGCGUA_AUAUAUCUAAUGUCCGGGGUCGUGGG__ .............................((((...........))))..(((((((((.((.................................)).))))))))).. (-13.63 = -12.52 + -1.11)

| Location | 17,123,643 – 17,123,736 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 60.69 |
| Shannon entropy | 0.76741 |
| G+C content | 0.50536 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
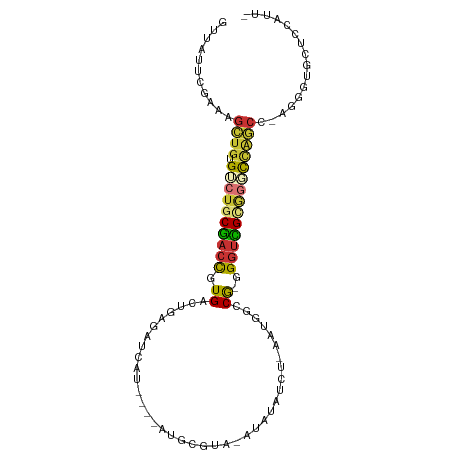
>dm3.chr3R 17123643 93 + 27905053 GUUAUUCGAAAGCUGUGUCUGCGACCGUGACUGAGAUCAU----AUGCGUACAUAUAUCU-AAUGUCCG-GGGUCGUAGGCCAGCC-AGGGUGCUCGAUU- ..(((((....((((.((((((((((.((((..((((.((----(((....)))))))))-...)).))-.)))))))))))))).-.))))).......- ( -35.90, z-score = -2.93, R) >droSec1.super_38 395266 93 - 400794 GUUAUUCGAAAGCUGUGUCUGCGACCAUGACUGAGAUUAU----AUGCGUACAUAUAUCU-AAUGUCCG-GGGUCGUGGGCCAGCC-AGGGCGCUCGAUU- .....((((..((((.(((..(((((..(((..((((.((----(((....)))))))))-...)))..-.)))))..)))))((.-...))))))))..- ( -34.20, z-score = -2.24, R) >droYak2.chr3R 5924023 93 + 28832112 GUUAUUCAAAAGCUGAGUCUGCGACCGUGACUGAGAUCAU----AUGCGUAUAUAUAUCU-AAUGUCCG-GGGUCGUAGGCCAGCC-AGGGUGCUCAAUU- ..(((((....((((.((((((((((.((((..((((.((----(((....)))))))))-...)).))-.)))))))))))))).-.))))).......- ( -32.70, z-score = -2.81, R) >droEre2.scaffold_4770 13209494 93 - 17746568 GUUAUUCGAAUGCUGAGUCUGCGACCGUGACUGAGAUCAU----AUGCGUAUAUAUAUCU-AAUGUCCG-GGGUCGUGGGCCAGCC-AGGGUACUCGAUU- ..(((((....((((.(((..(((((.((((..((((.((----(((....)))))))))-...)).))-.)))))..))))))).-.))))).......- ( -32.90, z-score = -1.89, R) >droAna3.scaffold_13340 3784271 101 + 23697760 GUUUGGUGAAAGUUGUGCCCACGACCGUGACAGCGAUAUAUAUAAUGUAAAAAUAUAUUUCGAUGGUCGUGGGUUGCAUAUCGACCCAGGGUCCCUCGAAU ((((((.(...((...((((((((((((.....(((.((((((.........)))))).))))))))))))))).)).....((((...))))).)))))) ( -32.00, z-score = -2.24, R) >dp4.chr2 5623962 82 + 30794189 -GUACGCGAAAGUUGUGGCUGCGACCCUGACUCAGCCUUA----GUGU-----------U-GAUUGUCG-GGGUCGCGGCCUGACC-AGGGUGCGUCGUUU -((((.(....((((.((((((((((((((((((((....----..))-----------)-))..))))-))))))))))))))).-.).))))....... ( -43.70, z-score = -4.92, R) >droPer1.super_19 1340722 82 + 1869541 -GUACGCGAAAGUUGUGGCUGCGACCCUGACUCAGCCUUA----GUGU-----------U-GAUUGUCG-GGGUCGCGGACUGACC-AGGGUGCGUCGUUU -(.(((((.......(((((((((((((((((((((....----..))-----------)-))..))))-))))))))).)))...-....)))))).... ( -37.34, z-score = -3.13, R) >droWil1.scaffold_181130 16102543 78 + 16660200 ------------GUGUGGUU-CUAGUUUGCCAGCGAUGAUGGUAUUGUUAA----UAUUAUCGUUGUCAUGGGCGGCGUGC------AGGUUACAUCAUUU ------------(((((...-((.((.((((((((((((((.((....)).----))))))))))((.....)))))).))------.)).)))))..... ( -20.20, z-score = -0.76, R) >consensus GUUAUUCGAAAGCUGUGUCUGCGACCGUGACUGAGAUCAU____AUGCGUA_AUAUAUCU_AAUGGCCG_GGGUCGCGGGCCAGCC_AGGGUGCUCCAUU_ ...........((((.((((((((((.............................................))))))))))))))................ (-14.27 = -14.27 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:56 2011