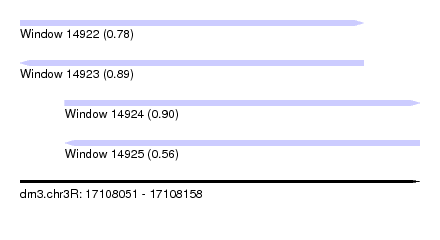
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,108,051 – 17,108,158 |
| Length | 107 |
| Max. P | 0.897950 |

| Location | 17,108,051 – 17,108,143 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.69 |
| Shannon entropy | 0.56854 |
| G+C content | 0.34009 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -8.72 |
| Energy contribution | -9.28 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778741 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 17108051 92 + 27905053 -----------------------CUCUAAGAC---CGUGGAGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGAUGG-CUCGUAAAAGGAGAGAAU- -----------------------(((......---......((((((((((.....(((((....)))))...))))))))))...((..(((((((...-.)))))))..)))))...- ( -19.50, z-score = -1.13, R) >droSim1.chr3R 23166118 92 - 27517382 -----------------------CCUUAAGAC---CGUGGAGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGAUGG-CUCGUAAAAAGAGAGAAC- -----------------------.........---......((((((((((.....(((((....)))))...))))))))))..(((..(((((((...-.)))))))..))).....- ( -16.80, z-score = -0.68, R) >droSec1.super_38 379581 92 - 400794 -----------------------CCCUAAGAC---CGUGGAGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGAUGG-CUCGUAAAAGGAGAGAAC- -----------------------.((......---...)).((((((((((.....(((((....)))))...))))))))))..(((..(((((((...-.)))))))..))).....- ( -18.10, z-score = -0.85, R) >droYak2.chr3R 5901770 92 + 28832112 -----------------------CCCAAAGUC---UGUGGGGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGAUGG-CUCGUAAAAGGAGAAAAC- -----------------------((((.....---..))))((((((((((.....(((((....)))))...))))))))))..(((..(((((((...-.)))))))..))).....- ( -22.00, z-score = -2.30, R) >droEre2.scaffold_4770 13193856 91 - 17746568 ------------------------CCUUAGUC---CAUGAAGAGAUUUAAGUUUCAUCUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGAUGG-CUCGUAAAAGGAGAAAAC- ------------------------......((---(.(((.(((((((((((((.(((.....)))...))).))))))))))...))).(((((((...-.))))))).)))......- ( -18.80, z-score = -1.79, R) >droAna3.scaffold_13340 3768508 117 + 23697760 UGGCAUUCAGCCCUUAAGCCCGCUUCUAAGACAA-CGAGUGGCGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCUAUUCGGUUUACGAGUG-CUCGUAAAAAUACAAAGC- .(((.....))).........((((.........-(((((((.((((((((.....(((((....)))))...)))))))).))))))).(((((((...-.)))))))......))))- ( -26.50, z-score = -1.76, R) >dp4.chr2 5608863 107 + 30794189 -----------ACUUAAGUCCGAGGCUAAGACAGCUGAGAAGAGAUUUAAGUUUCUUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGAGGG-CUCGUAAAAAUACAAAGC- -----------.........((((.((..(..(((((((..((((((((((....((((((....))))))..))))))))))..)))))))..)..)).-))))..............- ( -26.10, z-score = -2.23, R) >droPer1.super_19 1325481 107 + 1869541 -----------ACUUAAGUCCGAGGCUAAGACAGCUGAGAAGAGAUUUAAGUUUCUUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGAGUG-CUCGUAAAAAUACAAAGC- -----------.((((.((.....))))))...((((((..((((((((((....((((((....))))))..))))))))))..))))))((((((...-.))))))...........- ( -25.20, z-score = -2.02, R) >droWil1.scaffold_181130 16087081 99 + 16660200 ---------------------CCACAAGUCCAAACGAAAGAGAGAUUUAAGUUUCUUGUUUCAGAUAAUAAAUCUUAAAUUUCGAUUUGGUUUACGAGUGGCUCGUAAAAUAUAUACAAC ---------------------...((((((....(....).(((((((((((((.(((((...))))).))).)))))))))))))))).((((((((...))))))))........... ( -24.20, z-score = -3.48, R) >droVir3.scaffold_12822 2865433 76 - 4096053 ---------------------------------AUUGCGUGCUGAUUUCAGUUUCAGCUAGCAGAUAAUAAAUCUUAAAUUUCGCAUCACUUUACGAGUG-CUCGUAAAA---------- ---------------------------------..((((.(((((........)))))....((((.....)))).......))))....(((((((...-.))))))).---------- ( -17.50, z-score = -1.69, R) >droMoj3.scaffold_6540 297820 86 - 34148556 ---------------------------------AUUGCGUGCUGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAAUCGUUUUACGAGCG-CUCGUAAAAUACAAAGAAA ---------------------------------....((....((((((((.....(((((....)))))...)))))))).))....(((((((((...-.)))))))))......... ( -13.90, z-score = -0.05, R) >droGri2.scaffold_15074 2627304 85 + 7742996 ---------------------------------AGUGCGUGCUGAUUUAAGUU-CAUGUUUCGGAUAAUAAAUCUUAAAUUUGGAAUCACUUUACGAGUG-CUCGUAAAAUGGCAUGAAG ---------------------------------....(((((.........((-((.((((.((((.....)))).)))).))))..((.(((((((...-.))))))).)))))))... ( -19.40, z-score = -1.57, R) >consensus ________________________CCUAAGAC___UGUGGAGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGAGGG_CUCGUAAAAAGAGAAAAC_ .........................................(.((((((((.....(((((....)))))...)))))))).).......(((((((.....)))))))........... ( -8.72 = -9.28 + 0.56)
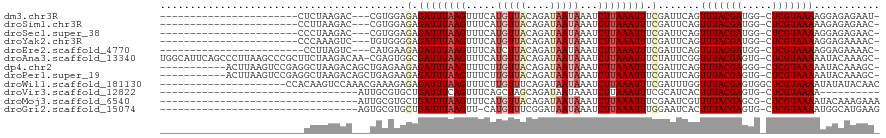


| Location | 17,108,051 – 17,108,143 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.69 |
| Shannon entropy | 0.56854 |
| G+C content | 0.34009 |
| Mean single sequence MFE | -17.28 |
| Consensus MFE | -8.82 |
| Energy contribution | -8.98 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891712 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 17108051 92 - 27905053 -AUUCUCUCCUUUUACGAG-CCAUCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCUCCACG---GUCUUAGAG----------------------- -...((((....((((((.-...))))))((((....((.(((((((.(((((..........))))).))))))).))....))---))...))))----------------------- ( -15.10, z-score = -1.46, R) >droSim1.chr3R 23166118 92 + 27517382 -GUUCUCUCUUUUUACGAG-CCAUCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCUCCACG---GUCUUAAGG----------------------- -......((..(((((((.-...)))))))..))...((.(((((((.(((((..........))))).))))))).))......---.........----------------------- ( -13.70, z-score = -0.89, R) >droSec1.super_38 379581 92 + 400794 -GUUCUCUCCUUUUACGAG-CCAUCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCUCCACG---GUCUUAGGG----------------------- -.......(((.((((((.-...))))))((((....((.(((((((.(((((..........))))).))))))).))....))---))...))).----------------------- ( -15.20, z-score = -0.97, R) >droYak2.chr3R 5901770 92 - 28832112 -GUUUUCUCCUUUUACGAG-CCAUCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCCCCACA---GACUUUGGG----------------------- -......((..(((((((.-...)))))))..))...((.(((((((.(((((..........))))).))))))).))((((..---.....))))----------------------- ( -15.70, z-score = -1.65, R) >droEre2.scaffold_4770 13193856 91 + 17746568 -GUUUUCUCCUUUUACGAG-CCAUCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAAGAUGAAACUUAAAUCUCUUCAUG---GACUAAGG------------------------ -.(((..(((.(((((((.-...))))))).((((..((.(((((((.((((((........)))))).))))))).)))))).)---))..))).------------------------ ( -18.10, z-score = -1.93, R) >droAna3.scaffold_13340 3768508 117 - 23697760 -GCUUUGUAUUUUUACGAG-CACUCGUAAACCGAAUAGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCGCCACUCG-UUGUCUUAGAAGCGGGCUUAAGGGCUGAAUGCCA -(((((.(((((((((((.-...))))))...))))).))).....((((((...((........))....))))))))..((((-((........))))))......(((.....))). ( -23.20, z-score = -0.14, R) >dp4.chr2 5608863 107 - 30794189 -GCUUUGUAUUUUUACGAG-CCCUCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAAGAAACUUAAAUCUCUUCUCAGCUGUCUUAGCCUCGGACUUAAGU----------- -((((.((...(((((((.-...)))))))((((...........(((((((...(((......)))....))))))).......((((...)))).))))))..))))----------- ( -17.40, z-score = -0.43, R) >droPer1.super_19 1325481 107 - 1869541 -GCUUUGUAUUUUUACGAG-CACUCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAAGAAACUUAAAUCUCUUCUCAGCUGUCUUAGCCUCGGACUUAAGU----------- -((((.((...(((((((.-...)))))))((((...........(((((((...(((......)))....))))))).......((((...)))).))))))..))))----------- ( -17.40, z-score = -0.25, R) >droWil1.scaffold_181130 16087081 99 - 16660200 GUUGUAUAUAUUUUACGAGCCACUCGUAAACCAAAUCGAAAUUUAAGAUUUAUUAUCUGAAACAAGAAACUUAAAUCUCUCUUUCGUUUGGACUUGUGG--------------------- ...........((((((((...))))))))(((((.(((((....(((((((...(((......)))....)))))))...))))))))))........--------------------- ( -22.20, z-score = -3.23, R) >droVir3.scaffold_12822 2865433 76 + 4096053 ----------UUUUACGAG-CACUCGUAAAGUGAUGCGAAAUUUAAGAUUUAUUAUCUGCUAGCUGAAACUGAAAUCAGCACGCAAU--------------------------------- ----------((((((((.-...))))))))...((((.......((((.....))))....(((((........))))).))))..--------------------------------- ( -19.80, z-score = -2.57, R) >droMoj3.scaffold_6540 297820 86 + 34148556 UUUCUUUGUAUUUUACGAG-CGCUCGUAAAACGAUUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCAGCACGCAAU--------------------------------- .....((((.((((((((.-...))))))))......(..(((((((.(((((..........))))).)))))))...)..)))).--------------------------------- ( -12.70, z-score = -0.34, R) >droGri2.scaffold_15074 2627304 85 - 7742996 CUUCAUGCCAUUUUACGAG-CACUCGUAAAGUGAUUCCAAAUUUAAGAUUUAUUAUCCGAAACAUG-AACUUAAAUCAGCACGCACU--------------------------------- .....(((((((((((((.-...)))))))))).......(((((((.(((((..........)))-)))))))))......)))..--------------------------------- ( -16.90, z-score = -3.20, R) >consensus _GUUUUCUCCUUUUACGAG_CCCUCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCUCCACA___GUCUUAGG________________________ ...........(((((((.....)))))))................((((((...((........))....))))))........................................... ( -8.82 = -8.98 + 0.17)


| Location | 17,108,063 – 17,108,158 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.30 |
| Shannon entropy | 0.48973 |
| G+C content | 0.32541 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -9.12 |
| Energy contribution | -9.34 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.897950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17108063 95 + 27905053 -----------GGAGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGA-UGGCUCGUAAAAGGAGA-GAAUAACAGCAAAACAACU------------ -----------...((((((((((.....(((((....)))))...))))))))))..(((..(((((((-....)))))))..))).-...................------------ ( -17.80, z-score = -1.77, R) >droSim1.chr3R 23166130 95 - 27517382 -----------GGAGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGA-UGGCUCGUAAAAAGAGA-GAACAACAGCAAAACAACU------------ -----------...((((((((((.....(((((....)))))...))))))))))..(((..(((((((-....)))))))..))).-...................------------ ( -16.80, z-score = -1.52, R) >droSec1.super_38 379593 95 - 400794 -----------GGAGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGA-UGGCUCGUAAAAGGAGA-GAACAACAGCAAAACAACU------------ -----------...((((((((((.....(((((....)))))...))))))))))..(((..(((((((-....)))))))..))).-...................------------ ( -17.80, z-score = -1.79, R) >droYak2.chr3R 5901782 95 + 28832112 -----------GGGGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGA-UGGCUCGUAAAAGGAGA-AAACCCCAGCAAAACAACU------------ -----------(((((((((((((.....(((((....)))))...))))))))))..(((..(((((((-....)))))))..))).-....)))............------------ ( -22.90, z-score = -2.92, R) >droEre2.scaffold_4770 13193867 95 - 17746568 -----------GAAGAGAUUUAAGUUUCAUCUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGA-UGGCUCGUAAAAGGAGA-AAACCACAGCAAAACAACU------------ -----------...(((((((((((((.(((.....)))...))).))))))))))..(((..(((((((-....)))))))..))).-...................------------ ( -17.70, z-score = -2.19, R) >droAna3.scaffold_13340 3768535 117 + 23697760 AAGACAAC-GAGUGGCGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCUAUUCGGUUUACGA-GUGCUCGUAAAAAUACA-AAGCAACAGCGGGUGCUGGCAAACAACAACU .......(-((((((.((((((((.....(((((....)))))...)))))))).)))))))((((.(.(-(..((((((....))).-..((....)))))..)).).))))....... ( -29.90, z-score = -3.04, R) >dp4.chr2 5608879 98 + 30794189 AAGACAGCUGAGAAGAGAUUUAAGUUUCUUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGA-GGGCUCGUAAAAAUACA-AAGCAGCGGAU-------------------- ......((((.((.((((((((((....((((((....))))))..))))))))))...))..(((((((-....)))))))......-...))))....-------------------- ( -22.90, z-score = -2.40, R) >droPer1.super_19 1325497 98 + 1869541 AAGACAGCUGAGAAGAGAUUUAAGUUUCUUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGA-GUGCUCGUAAAAAUACA-AAGCAGCGGAU-------------------- .....(((((((..((((((((((....((((((....))))))..))))))))))..)))))))..((.-.((((.(((....))).-.)))).))...-------------------- ( -23.00, z-score = -2.25, R) >droWil1.scaffold_181130 16087093 100 + 16660200 ------ACGAAAGAGAGAUUUAAGUUUCUUGUUUCAGAUAAUAAAUCUUAAAUUUCGAUUUGGUUUACGAGUGGCUCGUAAAAUAUAU-ACAACAAAGCAAAACAAC------------- ------.(.(((..(((((((((((((.(((((...))))).))).))))))))))..))).)((((((((...))))))))......-..................------------- ( -19.50, z-score = -2.04, R) >droMoj3.scaffold_6540 297825 93 - 34148556 -----------GUGCUGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAAUCGUUUUACGA-GCGCUCGUAAAAUACAAAGAAAUACAAAACAACU--------------- -----------((..((..(((((.....(((((....)))))...)))))(((((.....(((((((((-....))))))))).....))))).))..))....--------------- ( -14.60, z-score = -0.93, R) >droGri2.scaffold_15074 2627309 95 + 7742996 -----------GUGCUGAUUUAAGUU-CAUGUUUCGGAUAAUAAAUCUUAAAUUUGGAAUCACUUUACGA-GUGCUCGUAAAAUGGCAUGAAGUUCAAGAGCACAACA------------ -----------(((((..((.((.((-((((((((((((............)))))))..((.(((((((-....))))))).))))))))).)).)).)))))....------------ ( -25.10, z-score = -2.79, R) >consensus ___________GGAGAGAUUUAAGUUUCAUGUUACAGAUAAUAAAUCUUAAAUUUCGAUUCAGUUUACGA_UGGCUCGUAAAAAGAGA_AAACAACAGCAAAACAACU____________ ..............(.((((((((.....(((((....)))))...)))))))).).......(((((((.....)))))))...................................... ( -9.12 = -9.34 + 0.22)

| Location | 17,108,063 – 17,108,158 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.30 |
| Shannon entropy | 0.48973 |
| G+C content | 0.32541 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -7.88 |
| Energy contribution | -7.97 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17108063 95 - 27905053 ------------AGUUGUUUUGCUGUUAUUC-UCUCCUUUUACGAGCCA-UCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCUCC----------- ------------....((((((.((((((..-......(((((((....-))))))).((((((.........))))))......))))))))))))............----------- ( -15.40, z-score = -2.01, R) >droSim1.chr3R 23166130 95 + 27517382 ------------AGUUGUUUUGCUGUUGUUC-UCUCUUUUUACGAGCCA-UCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCUCC----------- ------------....((((((.((((((((-..((..(((((((....-)))))))..))...))).....((((.....)))).)))))))))))............----------- ( -14.90, z-score = -1.38, R) >droSec1.super_38 379593 95 + 400794 ------------AGUUGUUUUGCUGUUGUUC-UCUCCUUUUACGAGCCA-UCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCUCC----------- ------------....((((((.(((((...-......(((((((....-))))))).((((((.........)))))).......)))))))))))............----------- ( -14.20, z-score = -1.30, R) >droYak2.chr3R 5901782 95 - 28832112 ------------AGUUGUUUUGCUGGGGUUU-UCUCCUUUUACGAGCCA-UCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCCCC----------- ------------(((......)))((((.((-((((..(((((((....-)))))))..))...))))....(((((((...((........))....)))))))))))----------- ( -17.20, z-score = -1.04, R) >droEre2.scaffold_4770 13193867 95 + 17746568 ------------AGUUGUUUUGCUGUGGUUU-UCUCCUUUUACGAGCCA-UCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAAGAUGAAACUUAAAUCUCUUC----------- ------------......(((((.(((((((-...........))))))-).))))).......((.(((((((.((((((........)))))).))))))).))...----------- ( -17.90, z-score = -1.60, R) >droAna3.scaffold_13340 3768535 117 - 23697760 AGUUGUUGUUUGCCAGCACCCGCUGUUGCUU-UGUAUUUUUACGAGCAC-UCGUAAACCGAAUAGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCGCCACUC-GUUGUCUU .......((((((((((....)))).(((((-.(((....)))))))).-..))))))(((...(..(((((((.(((((..........))))).)))))))..)...))-)....... ( -21.60, z-score = -0.68, R) >dp4.chr2 5608879 98 - 30794189 --------------------AUCCGCUGCUU-UGUAUUUUUACGAGCCC-UCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAAGAAACUUAAAUCUCUUCUCAGCUGUCUU --------------------....((((.((-((.((((((((((....-))))))...)))))))).....(((((((...(((......)))....))))))).....))))...... ( -17.20, z-score = -2.05, R) >droPer1.super_19 1325497 98 - 1869541 --------------------AUCCGCUGCUU-UGUAUUUUUACGAGCAC-UCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAAGAAACUUAAAUCUCUUCUCAGCUGUCUU --------------------....((((.((-((.((((((((((....-))))))...)))))))).....(((((((...(((......)))....))))))).....))))...... ( -17.20, z-score = -1.66, R) >droWil1.scaffold_181130 16087093 100 - 16660200 -------------GUUGUUUUGCUUUGUUGU-AUAUAUUUUACGAGCCACUCGUAAACCAAAUCGAAAUUUAAGAUUUAUUAUCUGAAACAAGAAACUUAAAUCUCUCUUUCGU------ -------------.......(((......))-).....((((((((...))))))))......(((((....(((((((...(((......)))....)))))))...))))).------ ( -16.10, z-score = -1.53, R) >droMoj3.scaffold_6540 297825 93 + 34148556 ---------------AGUUGUUUUGUAUUUCUUUGUAUUUUACGAGCGC-UCGUAAAACGAUUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCAGCAC----------- ---------------.((((.((((..((((..(((.((((((((....-)))))))).(((...(((((...)))))...)))....))).))))..)))).))))..----------- ( -18.20, z-score = -1.42, R) >droGri2.scaffold_15074 2627309 95 - 7742996 ------------UGUUGUGCUCUUGAACUUCAUGCCAUUUUACGAGCAC-UCGUAAAGUGAUUCCAAAUUUAAGAUUUAUUAUCCGAAACAUG-AACUUAAAUCAGCAC----------- ------------....(((((....((.((((((.((((((((((....-))))))))))........(((..(((.....)))..)))))))-)).)).....)))))----------- ( -21.80, z-score = -3.07, R) >consensus ____________AGUUGUUUUGCUGUUGUUU_UCUCCUUUUACGAGCCA_UCGUAAACUGAAUCGAAAUUUAAGAUUUAUUAUCUGUAACAUGAAACUUAAAUCUCUCC___________ ......................................(((((((.....)))))))................((((((...((........))....))))))................ ( -7.88 = -7.97 + 0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:52 2011