| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,095,336 – 17,095,438 |
| Length | 102 |
| Max. P | 0.818330 |

| Location | 17,095,336 – 17,095,438 |
|---|---|
| Length | 102 |
| Sequences | 14 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.35 |
| Shannon entropy | 0.67079 |
| G+C content | 0.45735 |
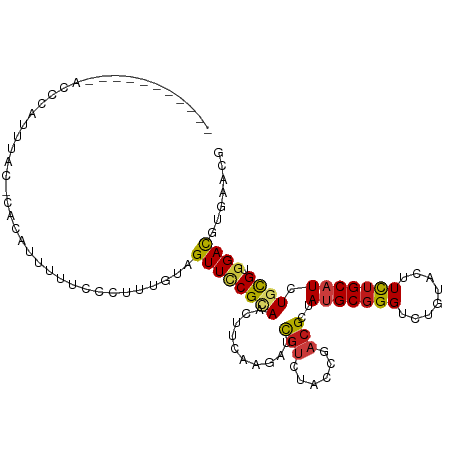
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -11.30 |
| Energy contribution | -11.09 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.818330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17095336 102 - 27905053 --------AUCACCCUUUUAU-CACACUUUUGCCUUUGUAGUUCCGCAACUUCAAGAUCGUCUACCGACGCUAUGCGGGUCUGUACUUCUGCAUCUGCGUGGACGUAAACG --------.............-......(((((.((((.((((....)))).))))...((((....((((.(((((((........)))))))..))))))))))))).. ( -21.70, z-score = -0.77, R) >droGri2.scaffold_15074 2613035 109 - 7742996 -AGUUAUAAACUAAUAGUUUC-CUUUCCUUUCUCACUCCAGUUUCGAAAUUUCAAAAUUGUAUAUAGACGCUAUGCGGGCCUCUACUUCUGCAUCUGCGUGGAUGUCAAUG -....................-...(((..........((((((.((....)).)))))).......((((.(((((((........)))))))..)))))))........ ( -18.10, z-score = -0.51, R) >droMoj3.scaffold_6540 285101 104 + 34148556 -------UUCUACUGACUAACGUACUGUGCAUCUUUUGCAGUUUCGUAAUUUCAAAAUUGUGUAUAGACGCUAUGCGGGUCUCUACUUUUGCAUUUGCGUGGACGUCAACG -------......((((.......((((((((.(((((.((((....)))).)))))..))))))))((((.(((((((........)))))))..))))....))))... ( -26.30, z-score = -1.70, R) >droVir3.scaffold_12822 2851067 97 + 4096053 -----------AUAUACUAAC-UGCUGUGCA--CUUCACAGUUUCGUAAUUUCAAAAUUGUGUACAGACGCUAUGCGGGUCUUUACUUUUGCAUCUGCGUGGACGUCAACG -----------..........-...((((((--(.........................)))))))(((((((((((((((.........).)))))))))).)))).... ( -25.41, z-score = -1.83, R) >droWil1.scaffold_181130 16074494 110 - 16660200 AUCCCUUCUAUUUUCAUUCUC-CCUACCACCCCCUCUUUAGUUCCGCAACUUUAAAAUUGUAUAUAGACGAUAUGCGGGCUUGUACUUCUGCAUCUGUGUGGAUGUUAACG .....................-....................((((((........(((((......)))))(((((((........)))))))...))))))........ ( -15.60, z-score = -0.20, R) >droPer1.super_19 1312776 94 - 1869541 ----------------UCUAA-CCUUUUACCCACUCCACAGUUUCGUAAUUUCAAAAUCGUCUACAGACGAUAUGCGGGUCUAUAUUUCUGCAUUUGCGUGGACGUCAAUG ----------------.....-..........(((((((.................((((((....))))))(((((((........)))))))....))))).))..... ( -21.80, z-score = -2.63, R) >dp4.chr2 5596177 94 - 30794189 ----------------UCUAA-CCUUUUACCCACUCCACAGUUUCGUAAUUUCAAAAUCGUCUACAGACGAUAUGCGGGUCUAUAUUUCUGCAUUUGCGUGGACGUCAAUG ----------------.....-..........(((((((.................((((((....))))))(((((((........)))))))....))))).))..... ( -21.80, z-score = -2.63, R) >droAna3.scaffold_13340 3756612 102 - 23697760 --------GGCACCCAUUUAA-UUACAAUCCCCACCCACAGUUCCGAAACUUCAAGAUCGUGUACCGAAGGUAUGCGGGCCUUUAUUUCUGCAUCUGCGUGGAUGUUAACG --------.............-..........((.((((......((((....(((.(((((((((...)))))))))..)))..)))).((....)))))).))...... ( -19.70, z-score = 0.54, R) >droEre2.scaffold_4770 13181458 102 + 17746568 --------AGAACCCCUUUAU-CUCAUUUUUGCCUUCGUAGUUCCGCAAUUUCAAGAUCGUCUACCGACGCUAUGCGGGUCUGUAUUUCUGCAUCUGCGUGGACGUGAACG --------.............-.((((..((((....)))).(((((............(((....)))((.(((((((........)))))))..))))))).))))... ( -21.30, z-score = 0.06, R) >droYak2.chr3R 5885690 101 - 28832112 --------UGAACCGCUUCAC-CACAUUUUU-CCUUCGUAGUUCCGCAAUUUCAAGAUCGUCUACCGACGCUAUGCGGGUCUGUAUUUCUGCAUCUGCGUGGAUGUGAACG --------.....((.(((((-.........-.((....)).(((((............(((....)))((.(((((((........)))))))..))))))).))))))) ( -23.20, z-score = -0.31, R) >droSec1.super_38 366834 102 + 400794 --------AGCACCCUUUUAU-CACAAUUUUGCCUUUGUAGUUCCGCAACUUCAAGAUCGUCUACCGACGCUAUGCGGGUCUGUAUUUCUGCAUCUGCGUGGACGUGAACG --------............(-(((.((((((...((((......))))...)))))).((((....((((.(((((((........)))))))..))))))))))))... ( -25.40, z-score = -1.13, R) >droSim1.chr3R 23153516 102 + 27517382 --------AGCACCCUUUUAU-CACAUUUUUGCCUUUGUAGUUCCGCAACUUCAAGAUCGUCUACCGACGCUAUGCGGGUCUGUAUUUCUGCAUCUGCGUGGACGUGAACG --------............(-(((..(((((...((((......))))...)))))..((((....((((.(((((((........)))))))..))))))))))))... ( -24.70, z-score = -0.94, R) >anoGam1.chr2R 8805827 87 - 62725911 -------------------AC-ACCAACUUUGUCG----AGUUCCGGAACUUCAAGAUCGUGUACCGCCGGUACGCGGGGCUGUACUUCUGCAUCUGCGUCGACGUGAACG -------------------..-.........((((----(((..(((((...((...(((((((((...)))))))))...))...))))).....)).)))))....... ( -23.90, z-score = 0.64, R) >triCas2.ChLG7 5329587 89 + 17478683 -----------------GAAU-GAUGCCUUU----UUGUAGUUCCGCAACUUCAAGAUUGUCUACCGUCGAUACGCCGGCCUCUACUUUUGCAUAUGCGUCGACGUGAACG -----------------....-...((..((----(((.((((....)))).)))))..))...(((((((..(((..((..........))....))))))))).).... ( -16.00, z-score = 0.72, R) >consensus ___________ACCCAUUUAC_CACAUUUUUCCCUUUGUAGUUCCGCAACUUCAAGAUCGUCUACCGACGCUAUGCGGGUCUGUACUUCUGCAUCUGCGUGGACGUGAACG ........................................((((((((..........(((......)))..(((((((........))))))).)))).))))....... (-11.30 = -11.09 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:49 2011