| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,091,686 – 17,091,838 |
| Length | 152 |
| Max. P | 0.988660 |

| Location | 17,091,686 – 17,091,838 |
|---|---|
| Length | 152 |
| Sequences | 8 |
| Columns | 177 |
| Reading direction | reverse |
| Mean pairwise identity | 75.35 |
| Shannon entropy | 0.45329 |
| G+C content | 0.38081 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -20.88 |
| Energy contribution | -20.70 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
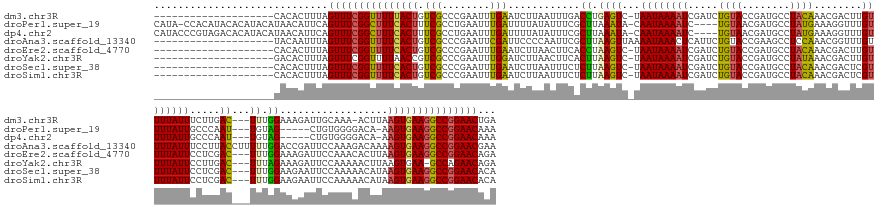
>dm3.chr3R 17091686 152 - 27905053 --------------------CACACUUUAGUUUCGGUUUUUACUGUCGCCCGAAUUUGAAUCUUAAUUUGACCUGAGUC-UAAUAAAAUCGAUCUGUACCGAUGCCUACAAACGACUUGUUUUAUUUCUUGAC---UUUGGAAAGAUUGCAAA-ACUUAAGUGAAGGCCGGAACUGA --------------------......((((((((((((((((((.((....)).((((((((((.......((.(((((-.((((((((((...((((........))))..)))....)))))))....)))---)).)).)))))).))))-.....)))))))))))))))))) ( -37.50, z-score = -2.83, R) >droPer1.super_19 1309084 162 - 1869541 CAUA-CCACAUACACAUACAUAACAUUCAGUUUCGGCUUUCACUUUCGCCUGAAUUUGAUUUUAUAUUUCGCUUAAAUA-CAAUAAAAUC----UGUAACGAUGCCUAUGAAAGGUUUGUUUUAUUGCCCAAU---UGUAG-----CUGUGGGGACA-AAGUGAAGGCCGGAACAAA ....-........................(((((((((((((((((..((((((......)))......(((.....((-((((......----...(((((.((((.....)))))))))..........))---)))).-----..))))))..)-))))))))))))))))... ( -40.81, z-score = -2.57, R) >dp4.chr2 5592483 163 - 30794189 CAUACCCGUAGACACAUACAUAACAUUCAGUUUCGGCUUUCACUUUCGCCUGAAUUUGAUUUUAUAUUUCGCUUAAAUA-CAAUAAAAUC----UGUAACGAUGCCUAUGAAAGGUUUGUUUUAUUGCCCAAU---UGUAG-----CUGUGGGGACA-AAGUGAAGGCCGGAACAAA .......(((......)))..........(((((((((((((((((..((((((......)))......(((.....((-((((......----...(((((.((((.....)))))))))..........))---)))).-----..))))))..)-))))))))))))))))... ( -42.01, z-score = -2.36, R) >droAna3.scaffold_13340 3753126 157 - 23697760 --------------------UACAAUUUAGUUUCGGUUUUCACUGUCGCCCGAAUUCGAUUCCCCAAUUCGCUUAAGUUAAAAUAAACUCAUUCUGUACCGAAGCCUCCAAACGGUUUGUUUUAUUUCCUUACCUUUUUGGACCGAUUCCAAAGACAAAAGUGAAGGCCGGAACGAA --------------------.........((((((((((((((((((...((((((.(.....).))))))..((((...(((((((..((..((((...((....))...))))..)).))))))).))))....((((((.....)))))))))...)))))))))))))))... ( -39.90, z-score = -2.69, R) >droEre2.scaffold_4770 13177893 153 + 17746568 --------------------CACACUUUAGUUUCGGUUUUCACUGUCGCCCGAAUUUGAAUCUUAACUUCACCUAAGUC-UAAUAAAAUCGAUCUGUACCGAUGCCUACAAACGACUUGUUUUAUUCCUCGAC---UUUGGAAAGAUUCCAAACACUUAAGUGAAGGCCGGAACAGA --------------------.........(((((((((((((((((((...((((.((((.......))))..((((((-.......((((........))))..........))))))....))))..))))---((((((.....))))))......)))))))))))))))... ( -39.03, z-score = -3.14, R) >droYak2.chr3R 5882103 152 - 28832112 --------------------GACACUUUAGUUUCGGUUUUAACCGUCGCCCGAAUUUGGAUCUUAACUUCACUUAAGUC-UAAUAAAAUCGAUCUGUACCGAUGCCUAUAAACGACUUGUUUUAUUCCUUGAC---UUUAGAAAGAUUCCAAAAACUUAAGUGAA-GCCAGAACAGA --------------------.........((..((((....))))..)).....((((..(((...((((((((((((.-......((((..((((.............((((.....))))...........---..))))..))))......)))))))))))-)..))).)))) ( -27.89, z-score = -0.70, R) >droSec1.super_38 363252 153 + 400794 --------------------CACACUUUAGUUUCGGUUUUCACUGUCGCCCGAAUUUGAAUCUUAAUUUCUCUUAAGUC-UAAUAAAAUCGAUCUGUACCGAUGCCUACAAACGACUCGUUUUAUUCCUCGAC---UUUGGAAGAAUUCCAAAAACAUAAGUGAAGGCCGGAACACA --------------------.........(((((((((((((((((((.............(((((......)))))..-.(((((((.(((((((((........))))...)).))))))))))...))))---((((((.....))))))......)))))))))))))))... ( -37.50, z-score = -2.92, R) >droSim1.chr3R 23149922 153 + 27517382 --------------------CACACUUUAGUUUCGGUUUUCACUGUCGCCCGAAUUUGAAUCUUAAUUUCUCUUAAGUC-UAAUAAAAUCGAUCUGUACCGAUGCCUACAAACGACUCGUUUUAUUCCUCGAC---UUUGGAAGAAUUCCAAAAACAUAAGUGAAGGCCGGAACACA --------------------.........(((((((((((((((((((.............(((((......)))))..-.(((((((.(((((((((........))))...)).))))))))))...))))---((((((.....))))))......)))))))))))))))... ( -37.50, z-score = -2.92, R) >consensus ____________________CACACUUUAGUUUCGGUUUUCACUGUCGCCCGAAUUUGAAUCUUAAUUUCACUUAAGUC_UAAUAAAAUCGAUCUGUACCGAUGCCUACAAACGACUUGUUUUAUUCCUCGAC___UUUGGAAAGAUUCCAAAAACAUAAGUGAAGGCCGGAACAAA .............................(((((((((((((((.(((........)))..............((((((........(((..........)))..........))))))........................................)))))))))))))))... (-20.88 = -20.70 + -0.18)
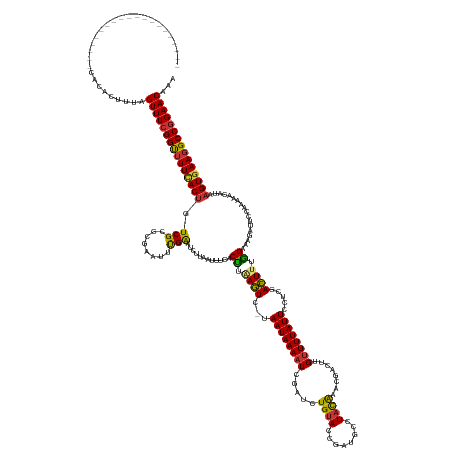
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:48 2011